#x ray crystallography
Photo


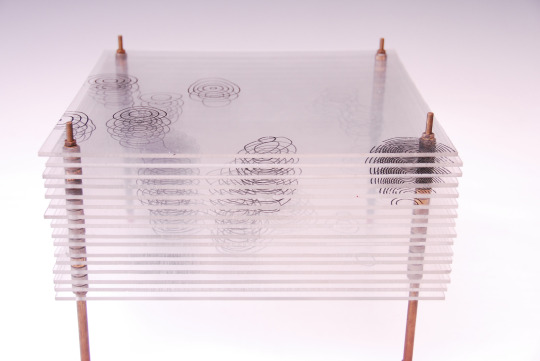
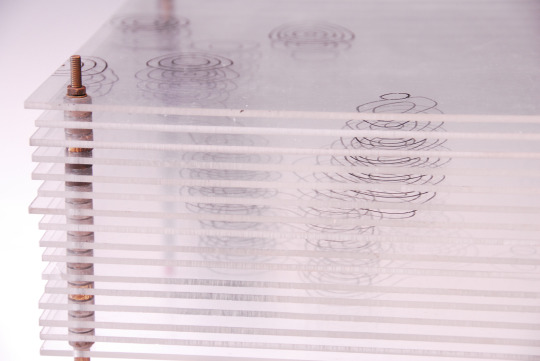
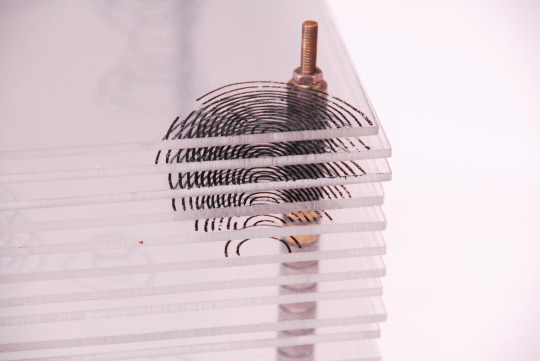



Model of the Structure of Penicillin, by Dorothy Crowfoot Hodgkin, Oxford, ca. 1945 [Based on X-ray crystallography work by Dorothy Crowfoot Hodgkin and Barbara Wharton Low (Oxford) and C. W. Bunn and A. Turner-Jones (I.C.I. Alkali Division, Northwich)] [© History of Science Museum, University of Oxford, Oxford]
#chemistry#x ray crystallography#crystallography#geometry#pattern#structure#dorothy crowfoot#dorothy crowfoot hodgkin#barbara wharton low#barbara low#c. w. bunn#a. turner jones#history of science museum#1940s
129 notes
·
View notes
Text
These are essentially iron-poryphyrin complexes, as shown in figure 13.48 on the next page.
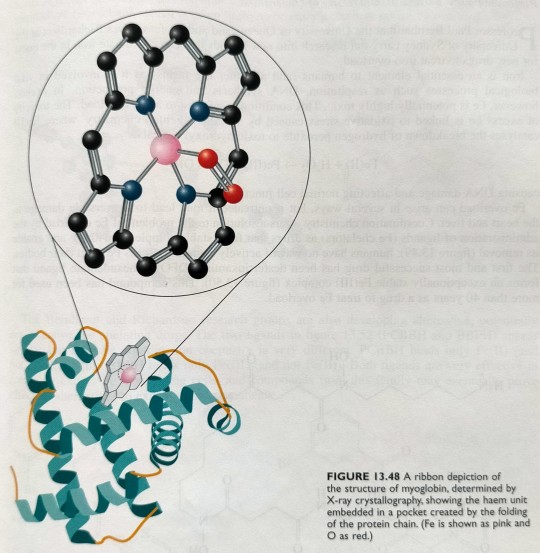
In myoglobin, the haem group is bound to a polypeptide chain of 153 amino acids arranged in helical arrays, and the ribbon structure of myoglobin is shown in figure 13.48.
"Chemistry" 2e - Blackman, A., Bottle, S., Schmid, S., Mocerino, M., Wille, U.
#book quotes#chemistry#nonfiction#textbook#ribbon#x ray crystallography#protein#amino acid#myoglobin#haem#iron#oxygen#nitrogen#helical
1 note
·
View note
Text
The New Zealand-born Rutherford was the first to show that the atom consists of a positively charged nucleus surrounded by tiny negatively charged electrons, while Bragg (born in Australia), together with his British-born father William Henry Bragg, developed the technique of X-ray crystallography, in which X-rays are used to determine the three-dimensional structure of solid matter on the atomic scale.
"Chemistry" 2e - Blackman, A., Bottle, S., Schmid, S., Mocerino, M., Wille, U.
#book quote#chemistry#nonfiction#textbook#australian#new zealander#ernest rutherford#william bragg#william lawrence bragg#william henry bragg#atom#nucleus#x ray#x ray crystallography#scientific discovery
0 notes
Link
Photosynthesis plays a crucial role in shaping and sustaining life on Earth, yet many aspects of the process remain a mystery. One such mystery is how Photosystem II, a protein complex in plants, algae and cyanobacteria, harvests energy from sunlight and uses it to split water, producing the oxygen we breathe. Now researchers from the Department of Energy’s SLAC National Accelerator Laboratory and Lawrence Berkeley National Laboratory, together with collaborators from Uppsala University, Humboldt University, and other institutions have succeeded in cracking a key secret of Photosystem II.
Using SLAC’s Linac Coherent Light Source (LCLS) and the SPring-8 Angstrom Compact free electron LAser (SACLA) in Japan, they captured for the first time in atomic detail what happens in the final moments leading up to the release of breathable oxygen. The data reveal an intermediate reaction step that had not been observed before.
The results, published today in Nature, shed light on how nature has optimized photosynthesis and are helping scientists develop artificial photosynthetic systems that mimic photosynthesis to harvest natural sunlight to convert carbon dioxide into hydrogen and carbon-based fuels.
Continue Reading
328 notes
·
View notes
Text
Beyond the Microscope: Cryo-EM’s Impact on Accelerating Drug Discovery.
A collaboration between the U.S. National Committees on: Crystallography, Chemistry, CODATA & the Board on Chemical Sciences and Technology.
As part of Advancing Drug Discovery: A Webinar Series of the National Academies of Sciences, Engineering and Medicine, a session on Beyond the Microscope: Cryo-EM’s Impact on Accelerating Drug Discovery will be held on Thursday, November 16th, 2023 at noon (EDT). The 60-minute session will consist a presentation from Sandra Gabelli, Head of Structural biology at Merck & co and formally Executive Director of Protein and Structural Chemistry.
Over the past decade, single particle cryo electron microscopy (cryoEM) has transitioned from a niche technique to a powerful tool for structural biology. This is due, in part, to technological breakthroughs which have made it relatively routine to achieve near molecule formulations for protein targets of pharmacological interest in complex with drug candidates. The pharmaceutical industry has historically relied heavily on X-ray crystallography to enable structure-based drug design (SBDD); however, today cryoEM is playing an ever-increasing role in that process, especially for challenging samples like large multimeric complexes, proteins that are flexible, or proteins that are difficult to express or purify. Dr. Gabelli will describe the strategy and present examples were cryoEM has impacted the pipeline at Merck by enabling biology, guiding SBDD, driving protein engineering of biologics and impacting small molecule formulations with micro ED. Dr. Gabelli will also explore the potential offered by upcoming technological advancements of the technique.Register

#X-ray crystallography#structure-based drug design (SBDD)#cryoEM#technological advancements#biological sciences#molecule formulations#medical sciences#strutural biology#Chemical Sciences and Technology#pharmaceutical
0 notes
Photo

Max von Laue was born on October 9, 1879. A German physicist who received the Nobel Prize in Physics in 1914 for his discovery of the diffraction of X-rays by crystals. In addition to his scientific endeavors with contributions in optics, crystallography, quantum theory, superconductivity, and the theory of relativity, Laue had a number of administrative positions which advanced and guided German scientific research and development during four decades. A strong objector to Nazism, he was instrumental in re-establishing and organizing German science after World War II.
#max von laue#physics#x-rays#diffraction#optics#crystallography#quantum theory#superconductivity#relativity#nobel prize#nobel prize winners#science#science history#science birthdays#on this day#on this day in science history
0 notes
Text
0 notes
Text
At the centre of Rosalind Franklin’s tombstone in London’s Willesden Jewish Cemetery is the word “scientist”. This is followed by the inscription, “Her research and discoveries on viruses remain of lasting benefit to mankind.”
As one of the twentieth century’s pre-eminent scientists, Franklin’s work has benefited all of humanity. The one-hundredth anniversary of her birth this month is prompting much reflection on her career and research contributions, not least Franklin’s catalytic role in unravelling the structure of DNA.
. . .
But Franklin’s remarkable work on DNA amounts to a fraction of her record and legacy. She was a tireless investigator of nature’s secrets, and worked across biology, chemistry and physics, with a focus on research that mattered to society. She made important advances in the science of coal and carbon, and she became an expert in the study of viruses that cause plant and human diseases. In essence, it is because of Franklin, her collaborators and successors, that today’s researchers are able to use tools such as DNA sequencing and X-ray crystallography to investigate viruses such as SARS-CoV-2.
. . .
Franklin was an inveterate traveller on the global conference circuit and a collaborator with international partners. She won a rare grant (with Klug) from the US National Institutes of Health. She was a global connector in the booming early days of research into virus structures: an expert in pathogenic viruses who had gained an international reputation and cared deeply about putting her research to use.
It is a travesty that Franklin is mostly remembered for not receiving full credit for her contributions to the discovery of DNA’s structure. That part of Franklin’s life story must never be forgotten, but she was so much more than the “wronged heroine”, and it’s time to recognize her for the full breadth and depth of her research career.
#rosalind franklin#nature#article#science#research#dna#covid#biology#chemistry#physics#medicine#health#histroy#women's history#x ray
609 notes
·
View notes
Text


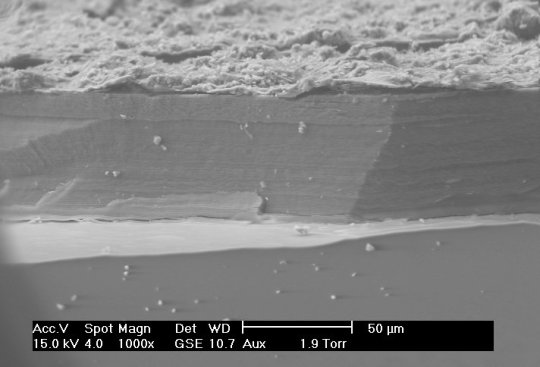
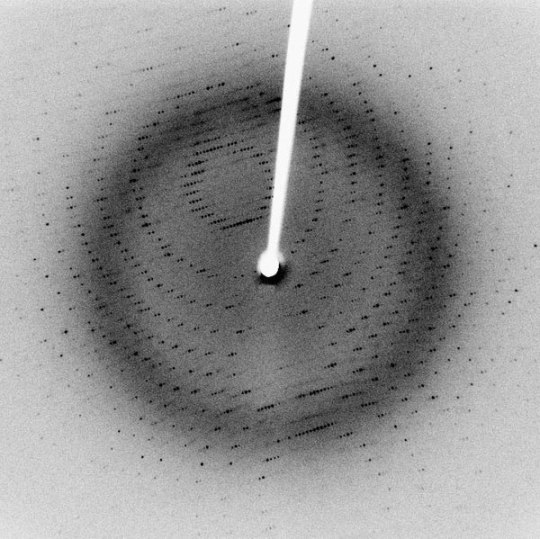
Small Scale Length Measurements
Many different techniques to measure length at small scales (typically microns to nanometers), including X-ray diffraction/crystallography, electron microscopy, and atomic force microscopy, among others. X-ray diffraction can be used to measure atomic spacing and lattice parameters (thereby determining crystal structure) using the wavelengths of the X-rays in question. Electron microscopy differs from many other techniques because they use, as the name implies, electrons to make their measurements, rather than any form of light. Though electrons are reflected back to a detector, the transit-time is not used to make measurements. Instead, Fourier transforms are used to generate an image.
Sources/Further Reading: (Images 1 and 4 - XRD Wikipedia) (Images 2 and 3 - SEM Wikipedia) (Measuring Length Wikipedia) (University of Wisconsin-Madison XRD) (University of Pennsylvania XRD) (University of Colorado XRD) (SEM Book Chapter)
#Materials Science#Science#Materials characterization#Electron microscopy#Diffraction#MeasurementMonday#2024Daily
12 notes
·
View notes
Text
19
The villain shoved the scientist into the work bench, glass vessels shattering on the floor as the scientist struggled to catch themself.
"You have twenty-four hours to identify that DNA sample," the villain told them, turning to leave. "Don't bother trying to escape. My men are surrounding the facility with orders to shoot to kill."
The scientist blinked. "Are you stupid?"
The villain's shoulders flinched, as if the scientist's words had physically whipped them. "Excuse me?" they growled, turning back around to face the scientist.
"Twenty-four hours is nowhere near enough time to identify a DNA sequence. Even the crystallization alone will take days. Do you even understand what you're demanding of me?"
"Do it faster then."
"I won't do it at all," the scientist spat.
Without a second thought, the villain took two steps forward and struck the scientist across the face. The scientist stumbled, mouth open in shock. Their cheek throbbed and they could feel the skin turning red.
"I don't think you understand the gravity of your situation," the villain said. They grabbed the scientist by the collar of their shirt and lifted, forcing the scientist to meet their eyes. "Your team is fully preoccupied with reconnaissance and I know you don't hire excess staff in the laboratory. The nearest city is an hour by car and I've seen to it that the nearest radio towers will not be receiving or transmitting any signals from this location. And did you not hear me when I said the building is surrounded? Do the words 'shoot to kill' ring a bell?"
The scientist fumed and silently cursed their remote location. Their intention to keep the lab a secret had certainly backfired.
They glared up at the villain and tried to steady their breathing. "The only way I can identify a DNA sequence is by x-ray crystallography. The crystallization process will take three to five days and the instrument takes around eighteen hours to analyze a sample. I need at least a day to recreate the sequence once I have the sample's results." They swallowed. "I need at least a week to do this for you. A quick internet search could have told you as much."
Still seething, the villain dropped the scientist and allowed them to straighten their shirt.
"Get started then," they bit out. "I'll be back in an hour with... revised instructions."
They left the lab and slammed the door behind them, the impact rattling a rack of test tubes. The scientist stared at the door for a minute, then, with a deep breath, took the broom and began sweeping up the broken glass.
#fiction#snippet#scientist#villain#scientist x villain#villain x scientist#writing#writeblr#annual post lol#no proofreading full send or nothing
82 notes
·
View notes
Photo

Dorothy Crowfoot Hodgkin, May 12, 1910 / 2022
(image: Dorothy Crowfoot Hodgkin assembling a molecular model. Photo: © Jorge Lewinski. Somerville College, University of Oxford, Oxford)
(plus: Dorothy Crowfoot Hodgkin, Nobel Lecture, December 11, 1964 (pdf here))
(plus: Guy Dodson, Dorothy Mary Crowfoot Hodgkin, O.M. 12 May 1910 – 29 July 1994, «Biographical Memoirs of Fellows of The Royal Society», Volume 48, The Royal Society, 2002, pp. 181-219 (pdf here))
(plus: Jenny P. Glusker, Dorothy Crowfoot Hodgkin (1910-1994), «Protein Science», 1994 Dec; 3(12): 2465–2469, Cambridge University Press, The Protein Society (pdf here))
#chemistry#x ray crystallography#crystallography#biology#photography#journal#dorothy crowfoot#dorothy crowfoot hodgkin#birthday anniversary#jorge lewinski#biographical memoirs of fellows of the royal society#protein science#the protein society
57 notes
·
View notes
Text
This arrangement of Na+ cations and Cl- anions was first determined in 1913 by Australian-born Sir William Lawrence Bragg, using X-ray equipment designed by his father, Sir William Henry Bragg (figure 5.10).


"Chemistry" 2e - Blackman, A., Bottle, S., Schmid, S., Mocerino, M., Wille, U.
#book quotes#chemistry#nonfiction#textbook#sodium#chlorine#sodium chloride#salt#cation#anion#ions#10s#1910s#20th century#william lawrence bragg#william henry bragg#x ray#x ray crystallography
0 notes
Text

Rosalind Franklin (1920-1958) was a British chemist and X-ray crystallographer whose work was crucial to the discovery of the double helix structure of DNA.
Here are some key facts about Rosalind Franklin:
She captured the famous "Photo 51" X-ray diffraction image of DNA in 1952, which revealed the double helix structure. However, her contribution was initially unrecognized.
She was an expert in X-ray crystallography and studied the molecular structures of DNA, viruses, coal and graphite.
Her work at King's College London, using X-ray diffraction techniques, provided important insights into the helical structure of DNA.
Her data and insights helped guide Francis Crick and James Watson in deducing the double helix model of DNA's 3D structure in 1953.
Franklin's contribution to the discovery was largely unacknowledged for many years due to the sexist attitudes of the time. Watson and Crick shared the 1962 Nobel Prize with Maurice Wilkins.
She made pioneering use of X-ray diffraction studies of viruses, laying the foundations for structural virology.
She died of ovarian cancer in 1958 at the age of 37, before she could be formally recognized for her seminal work on DNA structure.
Rosalind Franklin's meticulous X-ray work and skills in crystallography provided indispensable data that led to one of the greatest discoveries of the 20th century - the double helix structure of DNA.
#stickers#sticker art#sticker design#rosalind franklin#dna discovery#dna#biotechnology#science#sciart#scicomm
11 notes
·
View notes
Text
With that information, scientists could close in on the elusive mechanism of proton transfer, which could help to answer myriad questions in chemistry and biology. Knowing what protons are doing could have important implications in structural biology, where traditional methods like X-ray crystallography and cryo-electron microscopy have difficulty “seeing” protons.
18 notes
·
View notes
Text

The Nucleosome: DNA's Fancy Packaging and Party Trick!
Imagine cramming two meters of yarn into a pea-sized box. Sounds impossible, right? Well, that's the impressive feat that cells pull off every single day with DNA! They use a clever structure called the nucleosome to pack this massive genetic blueprint into the tiny nucleus.
The journey began in 1974 when Don and Ada Olins, peering through an electron microscope, spotted repeating beads – the first glimpse of nucleosomes. Roger Kornberg, building upon this observation, proposed the now-iconic "subunit theory," envisioning DNA wrapped around histone protein cores. This theory, later solidified by Pierre Oudet's term "nucleosome," laid the groundwork for further exploration. The 1980s witnessed a flurry of activity, with Aaron Klug's group using X-ray crystallography to reveal the left-handed superhelical twist of DNA around the histone octamer. But the true masterpiece arrived in 1997 when the Richmond group, armed with advanced techniques, unveiled the first near-atomic resolution crystal structure of the nucleosome. This intricate map, showcasing the precise interactions between DNA and histones, remains a cornerstone of our understanding.
The Players:
DNA: The star of the show, carrying our genetic code in the form of a double helix.
Histones: Protein spools around which DNA tightly winds. Imagine eight of them forming a core, like a mini-protein drum set.
Linker DNA: Short stretches of DNA connecting the spools, like the spaces between beads on a necklace.
The Steps:
Wrap and Roll: Picture DNA gracefully wrapping around the histone core, like thread around a spool. Each nucleosome holds about 146 base pairs of DNA, making about 1.67 turns.
Connect and Repeat: Linker DNA bridges the gap between nucleosomes, forming a "beads-on-a-string" structure. Think of it as pearls strung between the spools.
Compact and Condense: This repetitive unit folds further, creating intricate 30-nanometer fibers. Imagine these as twisted strands of pearls!
Here's the coolest part: histones aren't static. They can be chemically modified, like adding or removing phosphate groups. These modifications act like tiny flags that tell the cell how tightly to wrap the DNA, essentially throwing a "party" for specific genes by making them more accessible. This fine-tuning allows cells to respond to their environment and express the right genes at the right time. Understanding the nucleosome model is crucial for unraveling the mysteries of gene regulation and diseases like cancer. By studying how modifications affect nucleosome structure and gene access, scientists can develop new therapies to target specific genes and potentially treat diseases at the root cause.
While the nucleosome model is the foundation, the story gets even more intriguing. Different histone types and modifications create variations, influencing chromatin structure and function. Think of it as different music genres influencing the dance moves! Additionally, other proteins interact with the nucleosome, adding another layer of complexity to this fascinating choreography.
The nucleosome model is more than just a neat way to package DNA. It's a testament to the intricate dance between molecules that orchestrates life's processes. By understanding this fundamental structure, we gain deeper insights into cellular function, paving the way for advancements in medicine and beyond.
Remember, this is just the beginning! The world of nucleosomes and chromatin is vast and ever-evolving. So, keep exploring, keep questioning, and keep dancing to the rhythm of DNA!
#molecular biology#biology#science sculpt#life science#science#dna#biotechnology#genetics#Histone#Nucleosome#chromatin
7 notes
·
View notes
Text
Isabella Karle
youtube
Physical chemist Isabella Karle was born in 1921 in Detroit, Michigan. Karle is regarded as a pioneer in physical chemistry and crystallography. Her method for determining crystal structures formed the basis for all advanced x-ray crystallography, and dramatically improved both the accuracy and speed of chemical and biomedical analysis. Karle was the first to publish the structures of several complex organic and inorganic substances, and determined the structures of many important compounds. She was the author of more than 240 publications, and the winner of the 1995 National Medal of Science in Chemistry.
Isabella Karle died in 2017 at the age of 95.
11 notes
·
View notes