#Electrical Engineering&Computer Science (eecs)
Text
Three from MIT awarded 2024 Guggenheim Fellowships
New Post has been published on https://thedigitalinsider.com/three-from-mit-awarded-2024-guggenheim-fellowships/
Three from MIT awarded 2024 Guggenheim Fellowships


MIT faculty members Roger Levy, Tracy Slatyer, and Martin Wainwright are among 188 scientists, artists, and scholars awarded 2024 fellowships from the John Simon Guggenheim Memorial Foundation. Working across 52 disciplines, the fellows were selected from almost 3,000 applicants for “prior career achievement and exceptional promise.”
Each fellow receives a monetary stipend to pursue independent work at the highest level. Since its founding in 1925, the Guggenheim Foundation has awarded over $400 million in fellowships to more than 19,000 fellows. This year, MIT professors were recognized in the categories of neuroscience, physics, and data science.
Roger Levy is a professor in the Department of Brain and Cognitive Sciences. Combining computational modeling of large datasets with psycholinguistic experimentation, his work furthers our understanding of the cognitive underpinning of language processing, and helps to design models and algorithms that will allow machines to process human language. He is a recipient of the Alfred P. Sloan Research Fellowship, the NSF Faculty Early Career Development (CAREER) Award, and a fellowship at the Center for Advanced Study in the Behavioral Sciences.
Tracy Slatyer is a professor in the Department of Physics as well as the Center for Theoretical Physics in the MIT Laboratory for Nuclear Science and the MIT Kavli Institute for Astrophysics and Space Research. Her research focuses on dark matter — novel theoretical models, predicting observable signals, and analysis of astrophysical and cosmological datasets. She was a co-discoverer of the giant gamma-ray structures known as the “Fermi Bubbles” erupting from the center of the Milky Way, for which she received the New Horizons in Physics Prize in 2021. She is also a recipient of a Simons Investigator Award and Presidential Early Career Awards for Scientists and Engineers.
Martin Wainwright is the Cecil H. Green Professor in Electrical Engineering and Computer Science and Mathematics, and affiliated with the Laboratory for Information and Decision Systems and Statistics and Data Science Center. He is interested in statistics, machine learning, information theory, and optimization. Wainwright has been recognized with an Alfred P. Sloan Foundation Fellowship, the Medallion Lectureship and Award from the Institute of Mathematical Statistics, and the COPSS Presidents’ Award from the Joint Statistical Societies. Wainwright has also co-authored books on graphical and statistical modeling, and solo-authored a book on high dimensional statistics.
“Humanity faces some profound existential challenges,” says Edward Hirsch, president of the foundation. “The Guggenheim Fellowship is a life-changing recognition. It’s a celebrated investment into the lives and careers of distinguished artists, scholars, scientists, writers and other cultural visionaries who are meeting these challenges head-on and generating new possibilities and pathways across the broader culture as they do so.”
#000#2024#Algorithms#Analysis#artists#Astrophysics#Awards#honors and fellowships#book#Books#Brain#Brain and cognitive sciences#bubbles#career#career development#Careers#Center for Theoretical Physics#computer#Computer Science#Dark#dark matter#data#data science#datasets#Design#development#Electrical Engineering&Computer Science (eecs)#engineering#engineers#Faculty
0 notes
Photo

J-WAFS awards over $1.3 million in fourth round of seed grant funding Today, the Abdul Latif Jameel World Water and Food Security Lab (J-WAFS) at MIT announced the award of over $1.3 million in research funding through its seed grant program, now in its fourth year.
#Abdul Latif Jameel World Water and Food Security Lab (J-WAFS)#agriculture#Chemical engineering#Civil and environmental engineering#desalination#design#Developing countries#DMSE#EAPS#Electrical Engineering & Computer Science (eecs)#environment#food#Funding#Grants#IDSS#Invention#J-WAFS#Materials science and engineering#pollution#Research#School of Engineering#School of Science#Sensors#Sustainability#water
0 notes
Text
Two from MIT awarded 2024 Paul and Daisy Soros Fellowships for New Americans
New Post has been published on https://thedigitalinsider.com/two-from-mit-awarded-2024-paul-and-daisy-soros-fellowships-for-new-americans/
Two from MIT awarded 2024 Paul and Daisy Soros Fellowships for New Americans


MIT graduate student Riyam Al Msari and alumna Francisca Vasconcelos ’20 are among the 30 recipients of this year’s Paul and Daisy Soros Fellowships for New Americans. In addition, two Soros winners will begin PhD studies at MIT in the fall: Zijian (William) Niu in computational and systems biology and Russel Ly in economics.
The P.D. Soros Fellowships for New Americans program recognizes the potential of immigrants to make significant contributions to U.S. society, culture, and academia by providing $90,000 in graduate school financial support over two years.
Riyam Al Msari
Riyam Al Msari, born in Baghdad, Iraq, faced a turbulent childhood shaped by the 2003 war. At age 8, her life took a traumatic turn when her home was bombed in 2006, leading to her family’s displacement to Iraqi Kurdistan. Despite experiencing educational and ethnic discriminatory challenges, Al Msari remained undeterred, wholeheartedly embracing her education.
Soon after her father immigrated to the United States to seek political asylum in 2016, Al Msari’s mother was diagnosed with head and neck cancer, leaving Al Msari, at just 18, as her mother’s primary caregiver. Despite her mother’s survival, Al Msari witnessed the limitations and collateral damage caused by standardized cancer therapies, which left her mother in a compromised state. This realization invigorated her determination to pioneer translational cancer-targeted therapies.
In 2018, when Al Msari was 20, she came to the United States and reunited with her father and the rest of her family, who arrived later with significant help from then-senator Kamala Harris’s office. Despite her Iraqi university credits not transferring, Al Msari persevered and continued her education at Houston Community College as a Louis Stokes Alliances for Minority Participation (LSAMP) scholar, and then graduated magna cum laude as a Regents Scholar from the University of California at San Diego’s bioengineering program, where she focused on lymphatic-preserving neoadjuvant immunotherapies for head and neck cancers.
As a PhD student in the MIT Department of Biological Engineering, Al Masri conducts research in the Irvine and Wittrup labs to employ engineering strategies for localized immune targeting of cancers. She aspires to establish a startup that bridges preclinical and clinical oncology research, specializing in the development of innovative protein and biomaterial-based translational cancer immunotherapies.
Francisca Vasconcelos ’20
In the early 1990s, Francisca Vasconcelos’s parents emigrated from Portugal to the United States in pursuit of world-class scientific research opportunities. Vasconcelos was born in Boston while her parents were PhD students at MIT and Harvard University. When she was 5, her family relocated to San Diego, when her parents began working at the University of California at San Diego.
Vasconcelos graduated from MIT in 2020 with a BS in electrical engineering, computer science, and physics. As an undergraduate, she performed substantial research involving machine learning and data analysis for quantum computers in the MIT Engineering Quantum Systems Group, under the guidance of Professor William Oliver. Drawing upon her teaching and research experience at MIT, Vasconcelos became the founding academic director of The Coding School nonprofit’s Qubit x Qubit initiative, where she taught thousands of students from different backgrounds about the fundamentals of quantum computation.
In 2020, Vasconcelos was awarded a Rhodes Scholarship to the University of Oxford, where she pursued an MSc in statistical sciences and an MSt in philosophy of physics. At Oxford, she performed substantial research on uncertainty quantification of machine learning models for medical imaging in the OxCSML group. She also played for Oxford’s Women’s Blues Football team.
Now a computer science PhD student and NSF Graduate Research Fellow at the University of California at Berkeley, Vasconcelos is a member of both the Berkeley Artificial Intelligence Research Lab and CS Theory Group. Her research interests lie at the intersection of quantum computation and machine learning. She is especially interested in developing efficient classical algorithms to learn about quantum systems, as well as quantum algorithms to improve simulations of quantum processes. In doing so, she hopes to find meaningful ways in which quantum computers can outperform classical computers.
The P.D. Soros Fellowship attracts more than 1,800 applicants annually. MIT students interested in applying may contact Kim Benard, associate dean of distinguished fellowships in Career Advising and Professional Development.
#000#2024#Algorithms#Alumni/ae#Analysis#artificial#Artificial Intelligence#Awards#honors and fellowships#bioengineering#Biological engineering#Biology#Born#Cancer#career#Children#classical#coding#college#Community#computation#computer#Computer Science#computers#data#data analysis#development#Economics#education#Electrical Engineering&Computer Science (eecs)
0 notes
Text
Circadian rhythms can influence drugs’ effectiveness
New Post has been published on https://thedigitalinsider.com/circadian-rhythms-can-influence-drugs-effectiveness/
Circadian rhythms can influence drugs’ effectiveness


Giving drugs at different times of day could significantly affect how they are metabolized in the liver, according to a new study from MIT.
Using tiny, engineered livers derived from cells from human donors, the researchers found that many genes involved in drug metabolism are under circadian control. These circadian variations affect how much of a drug is available and how effectively the body can break it down. For example, they found that enzymes that break down Tylenol and other drugs are more abundant at certain times of day.
Overall, the researchers identified more than 300 liver genes that follow a circadian clock, including many involved in drug metabolism, as well as other functions such as inflammation. Analyzing these rhythms could help researchers develop better dosing schedules for existing drugs.
“One of the earliest applications for this method could be fine-tuning drug regimens of already approved drugs to maximize their efficacy and minimize their toxicity,” says Sangeeta Bhatia, the John and Dorothy Wilson Professor of Health Sciences and Technology and of Electrical Engineering and Computer Science at MIT, and a member of MIT’s Koch Institute for Integrative Cancer Research and the Institute for Medical Engineering and Science (IMES).
The study also revealed that the liver is more susceptible to infections such as malaria at certain points in the circadian cycle, when fewer inflammatory proteins are being produced.
Bhatia is the senior author of the new study, which appears today in Science Advances. The paper’s lead author is Sandra March, a research scientist in IMES.
Metabolic cycles
It is estimated that about 50 percent of human genes follow a circadian cycle, and many of these genes are active in the liver. However, exploring how circadian cycles affect liver function has been difficult because many of these genes are not identical in mice and humans, so mouse models can’t be used to study them.
Bhatia’s lab has previously developed a way to grow miniaturized livers using liver cells called hepatocytes, from human donors. In this study, she and her colleagues set out to investigate whether these engineered livers have their own circadian clocks.
Working with Charles Rice’s group at Rockefeller University, they identified culture conditions that support the circadian expression of a clock gene called Bmal1. This gene, which regulates the cyclic expression of a wide range of genes, allowed the liver cells to develop synchronized circadian oscillations. Then, the researchers measured gene expression in these cells every three hours for 48 hours, enabling them to identify more than 300 genes that were expressed in waves.
Most of these genes clustered in two groups — about 70 percent of the genes peaked together, while the remaining 30 percent were at their lowest point when the others peaked. These included genes involved in a variety of functions, including drug metabolism, glucose and lipid metabolism, and several immune processes.
Once the engineered livers established these circadian cycles, the researchers could use them to explore how circadian cycles affect liver function. First, they set out to study how time of day would affect drug metabolism, looking at two different drugs — acetaminophen (Tylenol) and atorvastatin, a drug used to treat high cholesterol.
When Tylenol is broken down in the liver, a small fraction of the drug is converted into a toxic byproduct known as NAPQI. The researchers found that the amount of NAPQI produced can vary by up to 50 percent, depending on what time of day the drug is administered. They also found that atorvastatin generates higher toxicity at certain times of day.
Both of these drugs are metabolized in part by an enzyme called CYP3A4, which has a circadian cycle. CYP3A4 is involved in processing about 50 percent of all drugs, so the researchers now plan to test more of those drugs using their liver models.
“In this set of drugs, it will be helpful to identify the time of the day to administer the drug to reach the highest effectiveness of the drug and minimize the adverse effects,” March says.
The MIT researchers are now working with collaborators to analyze a cancer drug they suspect may be affected by circadian cycles, and they hope to investigate whether this may also be true of drugs used in pain management.
Susceptibility to infection
Many of the liver genes that show circadian behavior are involved in immune responses such as inflammation, so the researchers wondered if this variation might influence susceptibility to infection. To answer that question, they exposed the engineered livers to Plasmodium falciparum, a parasite that causes malaria, at different points in the circadian cycle.
These studies revealed that the livers were more likely to become infected after exposure at different times of day. This is due to variations in the expression of genes called interferon-stimulated genes, which help to suppress infections.
“The inflammatory signals are much stronger at certain times of days than others,” Bhatia says. “This means that a virus like hepatitis or parasite like the one that causes malaria might be better at taking hold in your liver at certain times of the day.”
The researchers believe this cyclical variation may occur because the liver dampens its response to pathogens following meals, when it is typically exposed to an influx of microorganisms that might trigger inflammation even if they are not actually harmful.
Bhatia’s lab is now taking advantage of these cycles to study infections that are usually difficult to establish in engineered livers, including malaria infections caused by parasites other than Plasmodium falciparum.
“This is quite important for the field, because just by setting up the system and choosing the right time of infection, we can increase the infection rate of our culture by 25 percent, enabling drug screens that were otherwise impractical,” March says.
The research was funded by the MIT International Science and Technology Initiatives MIT-France program, the Koch Institute Support (core) Grant from the U.S. National Cancer Institute, the National Institute of Health and Medical Research of France, and the French National Research Agency.
#applications#Behavior#Cancer#Cells#circadian clock#circadian cycle#computer#Computer Science#drug#drugs#effects#Electrical Engineering&Computer Science (eecs)#engineering#enzyme#enzymes#Fraction#France#gene expression#genes#Giving#glucose#Health#Health sciences and technology#hepatitis#how#human#humans#infection#infections#inflammation
0 notes
Text
MIT scientists tune the entanglement structure in an array of qubits
New Post has been published on https://thedigitalinsider.com/mit-scientists-tune-the-entanglement-structure-in-an-array-of-qubits/
MIT scientists tune the entanglement structure in an array of qubits
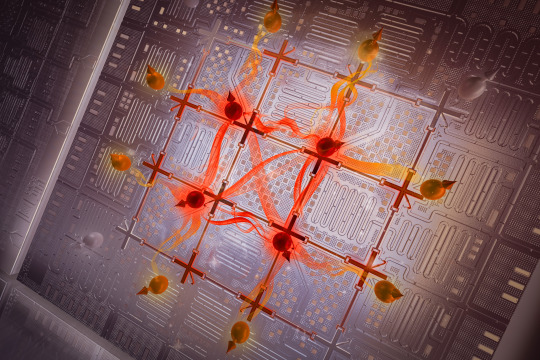

Entanglement is a form of correlation between quantum objects, such as particles at the atomic scale. This uniquely quantum phenomenon cannot be explained by the laws of classical physics, yet it is one of the properties that explains the macroscopic behavior of quantum systems.
Because entanglement is central to the way quantum systems work, understanding it better could give scientists a deeper sense of how information is stored and processed efficiently in such systems.
Qubits, or quantum bits, are the building blocks of a quantum computer. However, it is extremely difficult to make specific entangled states in many-qubit systems, let alone investigate them. There are also a variety of entangled states, and telling them apart can be challenging.
Now, MIT researchers have demonstrated a technique to efficiently generate entanglement among an array of superconducting qubits that exhibit a specific type of behavior.
Over the past years, the researchers at the Engineering Quantum Systems (EQuS) group have developed techniques using microwave technology to precisely control a quantum processor composed of superconducting circuits. In addition to these control techniques, the methods introduced in this work enable the processor to efficiently generate highly entangled states and shift those states from one type of entanglement to another — including between types that are more likely to support quantum speed-up and those that are not.
“Here, we are demonstrating that we can utilize the emerging quantum processors as a tool to further our understanding of physics. While everything we did in this experiment was on a scale which can still be simulated on a classical computer, we have a good roadmap for scaling this technology and methodology beyond the reach of classical computing,” says Amir H. Karamlou ’18, MEng ’18, PhD ’23, the lead author of the paper.
The senior author is William D. Oliver, the Henry Ellis Warren professor of electrical engineering and computer science and of physics, director of the Center for Quantum Engineering, leader of the EQuS group, and associate director of the Research Laboratory of Electronics. Karamlou and Oliver are joined by Research Scientist Jeff Grover, postdoc Ilan Rosen, and others in the departments of Electrical Engineering and Computer Science and of Physics at MIT, at MIT Lincoln Laboratory, and at Wellesley College and the University of Maryland. The research appears today in Nature.
Assessing entanglement
In a large quantum system comprising many interconnected qubits, one can think about entanglement as the amount of quantum information shared between a given subsystem of qubits and the rest of the larger system.
The entanglement within a quantum system can be categorized as area-law or volume-law, based on how this shared information scales with the geometry of subsystems. In volume-law entanglement, the amount of entanglement between a subsystem of qubits and the rest of the system grows proportionally with the total size of the subsystem.
On the other hand, area-law entanglement depends on how many shared connections exist between a subsystem of qubits and the larger system. As the subsystem expands, the amount of entanglement only grows along the boundary between the subsystem and the larger system.
In theory, the formation of volume-law entanglement is related to what makes quantum computing so powerful.
“While have not yet fully abstracted the role that entanglement plays in quantum algorithms, we do know that generating volume-law entanglement is a key ingredient to realizing a quantum advantage,” says Oliver.
However, volume-law entanglement is also more complex than area-law entanglement and practically prohibitive at scale to simulate using a classical computer.
“As you increase the complexity of your quantum system, it becomes increasingly difficult to simulate it with conventional computers. If I am trying to fully keep track of a system with 80 qubits, for instance, then I would need to store more information than what we have stored throughout the history of humanity,” Karamlou says.
The researchers created a quantum processor and control protocol that enable them to efficiently generate and probe both types of entanglement.
Their processor comprises superconducting circuits, which are used to engineer artificial atoms. The artificial atoms are utilized as qubits, which can be controlled and read out with high accuracy using microwave signals.
The device used for this experiment contained 16 qubits, arranged in a two-dimensional grid. The researchers carefully tuned the processor so all 16 qubits have the same transition frequency. Then, they applied an additional microwave drive to all of the qubits simultaneously.
If this microwave drive has the same frequency as the qubits, it generates quantum states that exhibit volume-law entanglement. However, as the microwave frequency increases or decreases, the qubits exhibit less volume-law entanglement, eventually crossing over to entangled states that increasingly follow an area-law scaling.
Careful control
“Our experiment is a tour de force of the capabilities of superconducting quantum processors. In one experiment, we operated the processor both as an analog simulation device, enabling us to efficiently prepare states with different entanglement structures, and as a digital computing device, needed to measure the ensuing entanglement scaling,” says Rosen.
To enable that control, the team put years of work into carefully building up the infrastructure around the quantum processor.
By demonstrating the crossover from volume-law to area-law entanglement, the researchers experimentally confirmed what theoretical studies had predicted. More importantly, this method can be used to determine whether the entanglement in a generic quantum processor is area-law or volume-law.
“The MIT experiment underscores the distinction between area-law and volume-law entanglement in two-dimensional quantum simulations using superconducting qubits. This beautifully complements our work on entanglement Hamiltonian tomography with trapped ions in a parallel publication published in Nature in 2023,” says Peter Zoller, a professor of theoretical physics at the University of Innsbruck, who was not involved with this work.
“Quantifying entanglement in large quantum systems is a challenging task for classical computers but a good example of where quantum simulation could help,” says Pedram Roushan of Google, who also was not involved in the study. “Using a 2D array of superconducting qubits, Karamlou and colleagues were able to measure entanglement entropy of various subsystems of various sizes. They measure the volume-law and area-law contributions to entropy, revealing crossover behavior as the system’s quantum state energy is tuned. It powerfully demonstrates the unique insights quantum simulators can offer.”
In the future, scientists could utilize this technique to study the thermodynamic behavior of complex quantum systems, which is too complex to be studied using current analytical methods and practically prohibitive to simulate on even the world’s most powerful supercomputers.
“The experiments we did in this work can be used to characterize or benchmark larger-scale quantum systems, and we may also learn something more about the nature of entanglement in these many-body systems,” says Karamlou.
Additional co-authors of the study are Sarah E. Muschinske, Cora N. Barrett, Agustin Di Paolo, Leon Ding, Patrick M. Harrington, Max Hays, Rabindra Das, David K. Kim, Bethany M. Niedzielski, Meghan Schuldt, Kyle Serniak, Mollie E. Schwartz, Jonilyn L. Yoder, Simon Gustavsson, and Yariv Yanay.
This research is funded, in part, by the U.S. Department of Energy, the U.S. Defense Advanced Research Projects Agency, the U.S. Army Research Office, the National Science Foundation, the STC Center for Integrated Quantum Materials, the Wellesley College Samuel and Hilda Levitt Fellowship, NASA, and the Oak Ridge Institute for Science and Education.
#2023#Algorithms#analog#artificial#atomic#atomic scale#atoms#Behavior#benchmark#Building#classical#college#complexity#computer#Computer Science#Computer science and technology#computers#computing#defense#Defense Advanced Research Projects Agency (DARPA)#Department of Energy (DoE)#education#Electrical Engineering&Computer Science (eecs)#Electronics#energy#Engineer#engineering#Explained#form#Foundation
0 notes
Text
Mapping the brain pathways of visual memorability
New Post has been published on https://thedigitalinsider.com/mapping-the-brain-pathways-of-visual-memorability/
Mapping the brain pathways of visual memorability


For nearly a decade, a team of MIT Computer Science and Artificial Intelligence Laboratory (CSAIL) researchers have been seeking to uncover why certain images persist in a people’s minds, while many others fade. To do this, they set out to map the spatio-temporal brain dynamics involved in recognizing a visual image. And now for the first time, scientists harnessed the combined strengths of magnetoencephalography (MEG), which captures the timing of brain activity, and functional magnetic resonance imaging (fMRI), which identifies active brain regions, to precisely determine when and where the brain processes a memorable image.
Their open-access study, published this month in PLOS Biology, used 78 pairs of images matched for the same concept but differing in their memorability scores — one was highly memorable and the other was easy to forget. These images were shown to 15 subjects, with scenes of skateboarding, animals in various environments, everyday objects like cups and chairs, natural landscapes like forests and beaches, urban scenes of streets and buildings, and faces displaying different expressions. What they found was that a more distributed network of brain regions than previously thought are actively involved in the encoding and retention processes that underpin memorability.
“People tend to remember some images better than others, even when they are conceptually similar, like different scenes of a person skateboarding,” says Benjamin Lahner, an MIT PhD student in electrical engineering and computer science, CSAIL affiliate, and first author of the study. “We’ve identified a brain signature of visual memorability that emerges around 300 milliseconds after seeing an image, involving areas across the ventral occipital cortex and temporal cortex, which processes information like color perception and object recognition. This signature indicates that highly memorable images prompt stronger and more sustained brain responses, especially in regions like the early visual cortex, which we previously underestimated in memory processing.”
While highly memorable images maintain a higher and more sustained response for about half a second, the response to less memorable images quickly diminishes. This insight, Lahner elaborated, could redefine our understanding of how memories form and persist. The team envisions this research holding potential for future clinical applications, particularly in early diagnosis and treatment of memory-related disorders.
The MEG/fMRI fusion method, developed in the lab of CSAIL Senior Research Scientist Aude Oliva, adeptly captures the brain’s spatial and temporal dynamics, overcoming the traditional constraints of either spatial or temporal specificity. The fusion method had a little help from its machine-learning friend, to better examine and compare the brain’s activity when looking at various images. They created a “representational matrix,” which is like a detailed chart, showing how similar neural responses are in various brain regions. This chart helped them identify the patterns of where and when the brain processes what we see.
Picking the conceptually similar image pairs with high and low memorability scores was the crucial ingredient to unlocking these insights into memorability. Lahner explained the process of aggregating behavioral data to assign memorability scores to images, where they curated a diverse set of high- and low-memorability images with balanced representation across different visual categories.
Despite strides made, the team notes a few limitations. While this work can identify brain regions showing significant memorability effects, it cannot elucidate the regions’ function in how it is contributing to better encoding/retrieval from memory.
“Understanding the neural underpinnings of memorability opens up exciting avenues for clinical advancements, particularly in diagnosing and treating memory-related disorders early on,” says Oliva. “The specific brain signatures we’ve identified for memorability could lead to early biomarkers for Alzheimer’s disease and other dementias. This research paves the way for novel intervention strategies that are finely tuned to the individual’s neural profile, potentially transforming the therapeutic landscape for memory impairments and significantly improving patient outcomes.”
“These findings are exciting because they give us insight into what is happening in the brain between seeing something and saving it into memory,” says Wilma Bainbridge, assistant professor of psychology at the University of Chicago, who was not involved in the study. “The researchers here are picking up on a cortical signal that reflects what’s important to remember, and what can be forgotten early on.”
Lahner and Oliva, who is also the director of strategic industry engagement at the MIT Schwarzman College of Computing, MIT director of the MIT-IBM Watson AI Lab, and CSAIL principal investigator, join Western University Assistant Professor Yalda Mohsenzadeh and York University researcher Caitlin Mullin on the paper. The team acknowledges a shared instrument grant from the National Institutes of Health, and their work was funded by the Vannevar Bush Faculty Fellowship via an Office of Naval Research grant, a National Science Foundation award, Multidisciplinary University Research Initiative award via an Army Research Office grant, and the EECS MathWorks Fellowship. Their paper is published in PLOS Biology.
#ai#Alzheimer's#Alzheimer's disease#Animals#applications#artificial#Artificial Intelligence#Biology#biomarkers#Brain#brain activity#Brain and cognitive sciences#buildings#chart#college#Color#computer#Computer Science#Computer Science and Artificial Intelligence Laboratory (CSAIL)#Computer science and technology#computing#data#Disease#disorders#dynamics#easy#effects#Electrical Engineering&Computer Science (eecs)#engineering#Explained
0 notes
Text
To build a better AI helper, start by modeling the irrational behavior of humans
New Post has been published on https://thedigitalinsider.com/to-build-a-better-ai-helper-start-by-modeling-the-irrational-behavior-of-humans/
To build a better AI helper, start by modeling the irrational behavior of humans


To build AI systems that can collaborate effectively with humans, it helps to have a good model of human behavior to start with. But humans tend to behave suboptimally when making decisions.
This irrationality, which is especially difficult to model, often boils down to computational constraints. A human can’t spend decades thinking about the ideal solution to a single problem.
Researchers at MIT and the University of Washington developed a way to model the behavior of an agent, whether human or machine, that accounts for the unknown computational constraints that may hamper the agent’s problem-solving abilities.
Their model can automatically infer an agent’s computational constraints by seeing just a few traces of their previous actions. The result, an agent’s so-called “inference budget,” can be used to predict that agent’s future behavior.
In a new paper, the researchers demonstrate how their method can be used to infer someone’s navigation goals from prior routes and to predict players’ subsequent moves in chess matches. Their technique matches or outperforms another popular method for modeling this type of decision-making.
Ultimately, this work could help scientists teach AI systems how humans behave, which could enable these systems to respond better to their human collaborators. Being able to understand a human’s behavior, and then to infer their goals from that behavior, could make an AI assistant much more useful, says Athul Paul Jacob, an electrical engineering and computer science (EECS) graduate student and lead author of a paper on this technique.
“If we know that a human is about to make a mistake, having seen how they have behaved before, the AI agent could step in and offer a better way to do it. Or the agent could adapt to the weaknesses that its human collaborators have. Being able to model human behavior is an important step toward building an AI agent that can actually help that human,” he says.
Jacob wrote the paper with Abhishek Gupta, assistant professor at the University of Washington, and senior author Jacob Andreas, associate professor in EECS and a member of the Computer Science and Artificial Intelligence Laboratory (CSAIL). The research will be presented at the International Conference on Learning Representations.
Modeling behavior
Researchers have been building computational models of human behavior for decades. Many prior approaches try to account for suboptimal decision-making by adding noise to the model. Instead of the agent always choosing the correct option, the model might have that agent make the correct choice 95 percent of the time.
However, these methods can fail to capture the fact that humans do not always behave suboptimally in the same way.
Others at MIT have also studied more effective ways to plan and infer goals in the face of suboptimal decision-making.
To build their model, Jacob and his collaborators drew inspiration from prior studies of chess players. They noticed that players took less time to think before acting when making simple moves and that stronger players tended to spend more time planning than weaker ones in challenging matches.
“At the end of the day, we saw that the depth of the planning, or how long someone thinks about the problem, is a really good proxy of how humans behave,” Jacob says.
They built a framework that could infer an agent’s depth of planning from prior actions and use that information to model the agent’s decision-making process.
The first step in their method involves running an algorithm for a set amount of time to solve the problem being studied. For instance, if they are studying a chess match, they might let the chess-playing algorithm run for a certain number of steps. At the end, the researchers can see the decisions the algorithm made at each step.
Their model compares these decisions to the behaviors of an agent solving the same problem. It will align the agent’s decisions with the algorithm’s decisions and identify the step where the agent stopped planning.
From this, the model can determine the agent’s inference budget, or how long that agent will plan for this problem. It can use the inference budget to predict how that agent would react when solving a similar problem.
An interpretable solution
This method can be very efficient because the researchers can access the full set of decisions made by the problem-solving algorithm without doing any extra work. This framework could also be applied to any problem that can be solved with a particular class of algorithms.
“For me, the most striking thing was the fact that this inference budget is very interpretable. It is saying tougher problems require more planning or being a strong player means planning for longer. When we first set out to do this, we didn’t think that our algorithm would be able to pick up on those behaviors naturally,” Jacob says.
The researchers tested their approach in three different modeling tasks: inferring navigation goals from previous routes, guessing someone’s communicative intent from their verbal cues, and predicting subsequent moves in human-human chess matches.
Their method either matched or outperformed a popular alternative in each experiment. Moreover, the researchers saw that their model of human behavior matched up well with measures of player skill (in chess matches) and task difficulty.
Moving forward, the researchers want to use this approach to model the planning process in other domains, such as reinforcement learning (a trial-and-error method commonly used in robotics). In the long run, they intend to keep building on this work toward the larger goal of developing more effective AI collaborators.
This work was supported, in part, by the MIT Schwarzman College of Computing Artificial Intelligence for Augmentation and Productivity program and the National Science Foundation.
#Accounts#agent#agents#ai#ai agent#AI AGENTS#ai assistant#AI systems#algorithm#Algorithms#approach#artificial#Artificial Intelligence#Behavior#Building#Capture#chess#collaborate#college#computer#Computer modeling#Computer Science#Computer Science and Artificial Intelligence Laboratory (CSAIL)#Computer science and technology#computing#conference#domains#Electrical Engineering&Computer Science (eecs)#engineering#Foundation
0 notes
Text
Four MIT faculty named 2023 AAAS Fellows
New Post has been published on https://thedigitalinsider.com/four-mit-faculty-named-2023-aaas-fellows/
Four MIT faculty named 2023 AAAS Fellows


Four MIT faculty members have been elected as fellows of the American Association for the Advancement of Science (AAAS).
The 2023 class of AAAS Fellows includes 502 scientists, engineers, and innovators across 24 scientific disciplines, who are being recognized for their scientifically and socially distinguished achievements.
Bevin Engelward initiated her scientific journey at Yale University under the mentorship of Thomas Steitz; following this, she pursued her doctoral studies at the Harvard School of Public Health under Leona Samson. In 1997, she became a faculty member at MIT, contributing to the establishment of the Department of Biological Engineering. Engelward’s research focuses on understanding DNA sequence rearrangements and developing innovative technologies for detecting genomic damage, all aimed at enhancing global public health initiatives.
William Oliver is the Henry Ellis Warren Professor of Electrical Engineering and Computer Science with a joint appointment in the Department of Physics, and was recently a Lincoln Laboratory Fellow. He serves as director of the Center for Quantum Engineering and associate director of the Research Laboratory of Electronics, and is a member of the National Quantum Initiative Advisory Committee. His research spans the materials growth, fabrication, 3D integration, design, control, and measurement of superconducting qubits and their use in small-scale quantum processors. He also develops cryogenic packaging and control electronics involving cryogenic complementary metal-oxide-semiconductors and single-flux quantum digital logic.
Daniel Rothman is a professor of geophysics in the Department of Earth, Atmospheric, and Planetary Sciences and co-director of the MIT Lorenz Center, a privately funded interdisciplinary research center devoted to learning how climate works. As a theoretical scientist, Rothman studies how the organization of the natural world emerges from the interactions of life and the physical environment. Using mathematics and statistical and nonlinear physics, he builds models that predict or explain observational data, contributing to our understanding of the dynamics of the carbon cycle and climate, instabilities and tipping points in the Earth system, and the dynamical organization of the microbial biosphere.
Vladan Vuletić is the Lester Wolfe Professor of Physics. His research areas include ultracold atoms, laser cooling, large-scale quantum entanglement, quantum optics, precision tests of physics beyond the Standard Model, and quantum simulation and computing with trapped neutral atoms. His Experimental Atomic Physics Group is also affiliated with the MIT-Harvard Center for Ultracold Atoms and the Research Laboratory of Electronics. In 2020, his group showed that the precision of current atomic clocks could be improved by entangling the atoms — a quantum phenomenon by which particles are coerced to behave in a collective, highly correlated state.
#2023#3d#atomic#atoms#Awards#honors and fellowships#Biological engineering#carbon#carbon cycle#climate#Collective#computer#Computer Science#computing#cooling#data#Design#DNA#dynamics#EAPS#earth#Electrical Engineering&Computer Science (eecs)#Electronics#engineering#engineers#Environment#experimental#Fabrication#Faculty#geophysics
0 notes
Text
For more open and equitable public discussions on social media, try “meronymity”
New Post has been published on https://thedigitalinsider.com/for-more-open-and-equitable-public-discussions-on-social-media-try-meronymity/
For more open and equitable public discussions on social media, try “meronymity”


Have you ever felt reluctant to share ideas during a meeting because you feared judgment from senior colleagues? You’re not alone. Research has shown this pervasive issue can lead to a lack of diversity in public discourse, especially when junior members of a community don’t speak up because they feel intimidated.
Anonymous communication can alleviate that fear and empower individuals to speak their minds, but anonymity also eliminates important social context and can quickly skew too far in the other direction, leading to toxic or hateful speech.
MIT researchers addressed these issues by designing a framework for identity disclosure in public conversations that falls somewhere in the middle, using a concept called “meronymity.”
Meronymity (from the Greek words for “partial” and “name”) allows people in a public discussion space to selectively reveal only relevant, verified aspects of their identity.
The researchers implemented meronymity in a communication system they built called LiTweeture, which is aimed at helping junior scholars use social media to ask research questions.
In LiTweeture, users can reveal a few professional facts, such as their academic affiliation or expertise in a certain field, which lends credibility to their questions or answers while shielding their exact identity.
Users have the flexibility to choose what they reveal about themselves each time they compose a social media post. They can also leverage existing relationships for endorsements that help queries reach experts they otherwise might be reluctant to contact.
During a monthlong study, junior academics who tested LiTweeture said meronymous communication made them feel more comfortable asking questions and encouraged them to engage with senior scholars on social media.
And while this study focused on academia, meronymous communication could be applied to any community or discussion space, says electrical engineering and computer science graduate student Nouran Soliman.
“With meronymity, we wanted to strike a balance between credibility and social inhibition. How can we make people feel more comfortable contributing and leveraging this rich community while still having some accountability?” says Soliman, lead author of a paper on meronymity.
Soliman wrote the paper with her advisor and senior author David Karger, professor in the Department of Electrical Engineering and Computer Science and a member of the Computer Science and Artificial Intelligence Laboratory (CSAIL), as well as others at the Semantic Scholar Team at Allen Institute for AI, the University of Washington, and Carnegie Mellon University. The research will be presented at the ACM Conference on Human Factors in Computing Systems.
Breaking down social barriers
The researchers began by conducting an initial study with 20 scholars to better understand the motivations and social barriers they face when engaging online with other academics.
They found that, while academics find X (formerly called Twitter) and Mastodon to be key resources when seeking help with research, they were often reluctant to ask for, discuss, or share recommendations.
Many respondents worried asking for help would make them appear to be unknowledgeable about a certain subject or feared public embarrassment if their posts were ignored.
The researchers developed LiTweeture to enable scholars to selectively present relevant facets of their identity when using social media to ask for research help.
But such identity markers, or “meronyms,” only give someone credibility if they are verified. So the researchers connected LiTweeture to Semantic Scholar, a web service which creates verified academic profiles for scholars detailing their education, affiliations, and publication history.
LiTweeture uses someone’s Semantic Scholar profile to automatically generate a set of meronyms they can choose to include with each social media post they compose. A meronym might be something like, “third-year graduate student at a research institution who has five publications at computer science conferences.”
A user writes a query and chooses the meronyms to appear with this specific post. LiTweeture then posts the query and meryonyms to X and Mastodon.
The user can also identify desired responders — perhaps certain researchers with relevant expertise — who will receive the query through a direct social media message or email. Users can personalize their meronyms for these experts, perhaps mentioning common colleagues or similar research projects.
Sharing social capital
They can also leverage connections by sharing their full identity with individuals who serve as public endorsers, such as an academic advisor or lab mate. Endorsements can encourage experts to respond to the asker’s query.
“The endorsement lets a senior figure donate some of their social capital to people who don’t have as much of it,” Karger says.
In addition, users can recruit close colleagues and peers to be helpers who are willing to repost their query so it reaches a wider audience.
Responders can answer queries using meronyms, which encourages potentially shy academics to offer their expertise, Soliman says.
The researchers tested LiTweeture during a field study with 13 junior academics who were tasked with writing and responding to queries. Participants said meronymous interactions gave them confidence when asking for help and provided high-quality recommendations.
Participants also used meronyms to seek a certain kind of answer. For instance, a user might disclose their publication history to signal that they are not seeking the most basic recommendations. When responding, individuals used identity signals to reflect their level of confidence in a recommendation, for example by disclosing their expertise.
“That implicit signaling was really interesting to see. I was also very excited to see that people wanted to connect with others based on their identity signals. This sense of relation also motivated some responders to make more effort when answering questions,” Soliman says.
Now that they have built a framework around academia, the researchers want to apply meronymity to other online communities and general social media conversations, especially those around issues where there is a lot of conflict, like politics. But to do that, they will need to find an effective, scalable way for people to present verified aspects of their identities.
“I think this is a tool that could be very helpful in many communities. But we have to figure out how to thread the needle on social inhibition. How can we create an environment where everyone feels safe speaking up, but also preserve enough accountability to discourage bad behavior? says Karger.
“Meronymity is not just a concept; it’s a novel technique that subtly blends aspects of identity and anonymity, creating a platform where credibility and privacy coexist. It changes digital communications by allowing safe engagement without full exposure, addressing the traditional anonymity-accountability trade-off. Its impact reaches beyond academia, fostering inclusivity and trust in digital interactions,” says Saiph Savage, assistant professor and director of the Civic A.I. Lab in the Khoury College of Computer Science at Northeastern University, and who was not involved with this work.
This research was funded, in part, by Semantic Scholar.
#ai#artificial#Artificial Intelligence#Behavior#Carnegie Mellon University#college#communication#communications#Community#computer#Computer Science#Computer Science and Artificial Intelligence Laboratory (CSAIL)#Computer science and technology#computing#computing systems#conference#Conflict#direction#diversity#education#Electrical Engineering&Computer Science (eecs)#email#engineering#Environment#Facts#fear#framework#Full#History#how
0 notes
Text
Women in STEM — A celebration of excellence and curiosity
New Post has been published on https://thedigitalinsider.com/women-in-stem-a-celebration-of-excellence-and-curiosity/
Women in STEM — A celebration of excellence and curiosity


What better way to commemorate Women’s History Month and International Women’s Day than to give three of the world’s most accomplished scientists an opportunity to talk about their careers? On March 7, MindHandHeart invited professors Paula Hammond, Ann Graybiel, and Sangeeta Bhatia to share their career journeys, from the progress they have witnessed to the challenges they have faced as women in STEM. Their conversation was moderated by Mary Fuller, chair of the faculty and professor of literature.
Hammond, an Institute professor with appointments in the Department of Chemical Engineering and the Koch Institute for Integrative Cancer Research, reflected on the strides made by women faculty at MIT, while acknowledging ongoing challenges. “I think that we have advanced a great deal in the last few decades in terms of the numbers of women who are present, although we still have a long way to go,” Hammond noted in her opening. “We’ve seen a remarkable increase over the past couple of decades in our undergraduate population here at MIT, and now we’re beginning to see it in the graduate population, which is really exciting.” Hammond was recently appointed to the role of vice provost for faculty.
Ann Graybiel, also an Institute professor, who has appointments in the Department of Brain and Cognitive Sciences and the McGovern Institute for Brain Research, described growing up in the Deep South. “Girls can’t do science,” she remembers being told in school, and they “can’t do research.” Yet her father, a physician scientist, often took her with him to work and had her assist from a young age, eventually encouraging her directly to pursue a career in science. Graybiel, who first came to MIT in 1973, noted that she continued to face barriers and rejection throughout her career long after leaving the South, but that individual gestures of inspiration, generosity, or simple statements of “You can do it” from her peers helped her power through and continue in her scientific pursuits.
Sangeeta Bhatia, the John and Dorothy Wilson Professor of Health Sciences and Technology and Electrical Engineering and Computer Science, director of the Marble Center for Cancer Nanomedicine at the Koch Institute for Integrative Cancer Research, and a member of the Institute for Medical Engineering and Science, is also the mother of two teenage girls. She shared her perspective on balancing career and family life: “I wanted to pick up my kids from school and I wanted to know their friends. … I had a vision for the life that I wanted.” Setting boundaries at work, she noted, empowered her to achieve both personal and professional goals. Bhatia also described her collaboration with President Emerita Susan Hockfield and MIT Amgen Professor of Biology Emerita Nancy Hopkins to spearhead the Future Founders Initiative, which aims to boost the representation of female faculty members pursuing biotechnology ventures.
A video of the full panel discussion is available on the MindHandHeart YouTube channel.
#Biology#biotechnology#Brain#Brain and cognitive sciences#brain research#Cancer#career#Careers#channel#chemical#Chemical engineering#Collaboration#computer#Computer Science#curiosity#deal#Diversity and Inclusion#Electrical Engineering&Computer Science (eecs)#engineering#Faculty#Full#Future#Health#Health sciences and technology#History#History of MIT#how#Inspiration#Institute for Medical Engineering and Science (IMES)#International Women's Day
0 notes
Text
A faster, better way to prevent an AI chatbot from giving toxic responses
New Post has been published on https://thedigitalinsider.com/a-faster-better-way-to-prevent-an-ai-chatbot-from-giving-toxic-responses/
A faster, better way to prevent an AI chatbot from giving toxic responses


A user could ask ChatGPT to write a computer program or summarize an article, and the AI chatbot would likely be able to generate useful code or write a cogent synopsis. However, someone could also ask for instructions to build a bomb, and the chatbot might be able to provide those, too.
To prevent this and other safety issues, companies that build large language models typically safeguard them using a process called red-teaming. Teams of human testers write prompts aimed at triggering unsafe or toxic text from the model being tested. These prompts are used to teach the chatbot to avoid such responses.
But this only works effectively if engineers know which toxic prompts to use. If human testers miss some prompts, which is likely given the number of possibilities, a chatbot regarded as safe might still be capable of generating unsafe answers.
Researchers from Improbable AI Lab at MIT and the MIT-IBM Watson AI Lab used machine learning to improve red-teaming. They developed a technique to train a red-team large language model to automatically generate diverse prompts that trigger a wider range of undesirable responses from the chatbot being tested.
They do this by teaching the red-team model to be curious when it writes prompts, and to focus on novel prompts that evoke toxic responses from the target model.
The technique outperformed human testers and other machine-learning approaches by generating more distinct prompts that elicited increasingly toxic responses. Not only does their method significantly improve the coverage of inputs being tested compared to other automated methods, but it can also draw out toxic responses from a chatbot that had safeguards built into it by human experts.
“Right now, every large language model has to undergo a very lengthy period of red-teaming to ensure its safety. That is not going to be sustainable if we want to update these models in rapidly changing environments. Our method provides a faster and more effective way to do this quality assurance,” says Zhang-Wei Hong, an electrical engineering and computer science (EECS) graduate student in the Improbable AI lab and lead author of a paper on this red-teaming approach.
Hong’s co-authors include EECS graduate students Idan Shenfield, Tsun-Hsuan Wang, and Yung-Sung Chuang; Aldo Pareja and Akash Srivastava, research scientists at the MIT-IBM Watson AI Lab; James Glass, senior research scientist and head of the Spoken Language Systems Group in the Computer Science and Artificial Intelligence Laboratory (CSAIL); and senior author Pulkit Agrawal, director of Improbable AI Lab and an assistant professor in CSAIL. The research will be presented at the International Conference on Learning Representations.
Automated red-teaming
Large language models, like those that power AI chatbots, are often trained by showing them enormous amounts of text from billions of public websites. So, not only can they learn to generate toxic words or describe illegal activities, the models could also leak personal information they may have picked up.
The tedious and costly nature of human red-teaming, which is often ineffective at generating a wide enough variety of prompts to fully safeguard a model, has encouraged researchers to automate the process using machine learning.
Such techniques often train a red-team model using reinforcement learning. This trial-and-error process rewards the red-team model for generating prompts that trigger toxic responses from the chatbot being tested.
But due to the way reinforcement learning works, the red-team model will often keep generating a few similar prompts that are highly toxic to maximize its reward.
For their reinforcement learning approach, the MIT researchers utilized a technique called curiosity-driven exploration. The red-team model is incentivized to be curious about the consequences of each prompt it generates, so it will try prompts with different words, sentence patterns, or meanings.
“If the red-team model has already seen a specific prompt, then reproducing it will not generate any curiosity in the red-team model, so it will be pushed to create new prompts,” Hong says.
During its training process, the red-team model generates a prompt and interacts with the chatbot. The chatbot responds, and a safety classifier rates the toxicity of its response, rewarding the red-team model based on that rating.
Rewarding curiosity
The red-team model’s objective is to maximize its reward by eliciting an even more toxic response with a novel prompt. The researchers enable curiosity in the red-team model by modifying the reward signal in the reinforcement learning set up.
First, in addition to maximizing toxicity, they include an entropy bonus that encourages the red-team model to be more random as it explores different prompts. Second, to make the agent curious they include two novelty rewards. One rewards the model based on the similarity of words in its prompts, and the other rewards the model based on semantic similarity. (Less similarity yields a higher reward.)
To prevent the red-team model from generating random, nonsensical text, which can trick the classifier into awarding a high toxicity score, the researchers also added a naturalistic language bonus to the training objective.
With these additions in place, the researchers compared the toxicity and diversity of responses their red-team model generated with other automated techniques. Their model outperformed the baselines on both metrics.
They also used their red-team model to test a chatbot that had been fine-tuned with human feedback so it would not give toxic replies. Their curiosity-driven approach was able to quickly produce 196 prompts that elicited toxic responses from this “safe” chatbot.
“We are seeing a surge of models, which is only expected to rise. Imagine thousands of models or even more and companies/labs pushing model updates frequently. These models are going to be an integral part of our lives and it’s important that they are verified before released for public consumption. Manual verification of models is simply not scalable, and our work is an attempt to reduce the human effort to ensure a safer and trustworthy AI future,” says Agrawal.
In the future, the researchers want to enable the red-team model to generate prompts about a wider variety of topics. They also want to explore the use of a large language model as the toxicity classifier. In this way, a user could train the toxicity classifier using a company policy document, for instance, so a red-team model could test a chatbot for company policy violations.
“If you are releasing a new AI model and are concerned about whether it will behave as expected, consider using curiosity-driven red-teaming,” says Agrawal.
This research is funded, in part, by Hyundai Motor Company, Quanta Computer Inc., the MIT-IBM Watson AI Lab, an Amazon Web Services MLRA research grant, the U.S. Army Research Office, the U.S. Defense Advanced Research Projects Agency Machine Common Sense Program, the U.S. Office of Naval Research, the U.S. Air Force Research Laboratory, and the U.S. Air Force Artificial Intelligence Accelerator.
#ai#AI Chatbot#ai model#air#air force#Algorithms#Amazon#Amazon Web Services#approach#Article#artificial#Artificial Intelligence#chatbot#chatbots#chatGPT#code#Companies#computer#Computer Science#Computer Science and Artificial Intelligence Laboratory (CSAIL)#Computer science and technology#conference#curiosity#defense#Defense Advanced Research Projects Agency (DARPA)#diversity#Electrical Engineering&Computer Science (eecs)#engineering#engineers#focus
0 notes
Text
For Julie Greenberg, a career of research, mentoring, and advocacy
New Post has been published on https://thedigitalinsider.com/for-julie-greenberg-a-career-of-research-mentoring-and-advocacy/
For Julie Greenberg, a career of research, mentoring, and advocacy


For Julie E. Greenberg SM ’89, PhD ’94, what began with a middle-of-the-night phone call from overseas became a gratifying career of study, research, mentoring, advocacy, and guiding of the office of a unique program with a mission to educate the next generation of clinician-scientists and engineers.
In 1987, Greenberg was a computer engineering graduate of the University of Michigan, living in Tel Aviv, Israel, where she was working for Motorola — when she answered an early-morning call from Roger Mark, then the director of the Harvard-MIT Program in Health Sciences and Technology (HST). A native of Detroit, Michigan, Greenberg had just been accepted into MIT’s electrical engineering and computer science (EECS) graduate program.
HST — one of the world’s oldest interdisciplinary educational programs based on translational medical science and engineering — had been offering the medical engineering and medical physics (MEMP) PhD program since 1978, but it was then still relatively unknown. Mark, an MIT distinguished professor of health sciences and technology and of EECS, and assistant professor of medicine at Harvard Medical School, was calling to ask Greenberg if she might be interested in enrolling in HST’s MEMP program.
“At the time, I had applied to MIT not knowing that HST existed,” Greenberg recalls. “So, I was groggily answering the phone in the middle of the night and trying to be quiet, because my roommate was a co-worker at Motorola, and no one yet knew that I was planning to leave to go to grad school. Roger asked if I’d like to be considered for HST, but he also suggested that I could come to EECS in the fall, learn more about HST, and then apply the following year. That was the option I chose.”
For Greenberg, who retired March 15 from her role as senior lecturer and director of education — that early morning phone call was the first she would hear of the program where she would eventually spend the bulk of her 37-year career at MIT, first as a student, then as the director of HST’s academic office. During her first year as a graduate student, she enrolled in class HST.582/6.555 (Biomedical Signal and Image Processing), for which she later served as lecturer and eventually course director, teaching the class almost every year for three decades. But as a first-year graduate student, she says she found that “all the cool kids” were HST students. “It was a small class, so we all got to know each other,” Greenberg remembers. “EECS was a big program. The MEMP students were a tight, close-knit community, so in addition to my desire to work on biomedical applications, that made HST very appealing.”
Also piquing her interest in HST was meeting Martha L. Gray, the Whitaker Professor in Biomedical Engineering. Gray, who is also a professor of EECS and a core faculty member of the MIT Institute for Medical Engineering and Science (IMES), was then a new member of the EECS faculty, and Greenberg met her at an orientation event for graduate student women, who were a smaller cohort then, compared to now. Gray SM ’81, PhD ’86 became Greenberg’s academic advisor when she joined HST. Greenberg’s SM and PhD research was on signal processing for hearing aids, in what was then the Sensory Communication Group in MIT’s Research Laboratory of Electronics (RLE).
Gray later succeeded Mark as director of HST at MIT, and it was she who recruited Greenberg to join as HST director of education in 2004, after Greenberg had spent a decade as a researcher in RLE.
“Julie is amazing — one of my best decisions as HST director was to hire Julie. She is an exceptionally clear thinker, a superb collaborator, and wicked smart,” Gray says. “One of her superpowers is being able to take something that is incredibly complex and to break it down into logical chunks … And she is absolutely devoted to advocating for the students. She is no pushover, but she has a way of coming up with solutions to what look like unfixable problems, before they become even bigger.”
Greenberg’s experience as an HST graduate student herself has informed her leadership, giving her a unique perspective on the challenges for those who are studying and researching in a demanding program that flows between two powerful institutions. HST students have full access to classes and all academic and other opportunities at both MIT and Harvard University, while having a primary institution for administrative purposes, and ultimately to award their degree. HST’s home at Harvard is in the London Society at Harvard Medical School, while at MIT, it is IMES.
In looking back on her career in HST, Greenberg says the overarching theme is one of “doing everything possible to smooth the path. So that students can get to where they need to go, and learn what they need to learn, and do what they need to do, rather than getting caught up in the bureaucratic obstacles of maneuvering between institutions. Having been through it myself gives me a good sense of how to empower the students.”
Rachel Frances Bellisle, an HST MEMP student who is graduating in May and is studying bioastronautics, says that having Julie as her academic advisor was invaluable because of her eagerness to solve the thorniest of issues. “Whenever I was trying to navigate something and was having trouble finding a solution, Julie was someone I could always turn to,” she says. “I know many graduate students in other programs who haven’t had the important benefit of that sort of individualized support. She’s always had my back.”
And Xining Gao, a fourth-year MEMP student studying biological engineering, says that as a student who started during the Covid pandemic, having someone like Greenberg and the others in the HST academic office — who worked to overcome the challenges of interacting mostly over Zoom — made a crucial difference. “A lot of us who joined in 2020 felt pretty disconnected,” Gao says. “Julie being our touchstone and guide in the absence of face-to-face interactions was so key.” The pandemic challenges inspired Gao to take on student government positions, including as PhD co-chair of the HST Joint Council. “Working with Julie, I’ve seen firsthand how committed she is to our department,” Gao says. “She is truly a cornerstone of the HST community.”
During her time at MIT, Greenberg has been involved in many Institute-level initiatives, including as a member of the 2016 class of the Leader to Leader program. She lauded L2L as being “transformative” to her professional development, saying that there have been “countless occasions where I’ve been able to solve a problem quickly and efficiently by reaching out to a fellow L2L alum in another part of the Institute.”
Since Greenberg started leading HST operations, the program has steadily evolved. When Greenberg was a student, the MEMP class was relatively small, admitting 10 students annually, with roughly 30 percent of them being women. Now, approximately 20 new MEMP PhD students and 30 new MD or MD-PhD students join the HST community each year, and half of them are women. Since 2004, the average time-to-degree for HST MEMP PhD students dropped by almost a full year, and is now on par with the average for all graduate programs in MIT’s School of Engineering, despite the complications of taking classes at both Harvard and MIT.
A search is underway for Julie’s replacement. But in the meantime, those who have worked with her praise her impact on HST, and on MIT.
“Throughout the entire history of the HST ecosystem, you cannot find anyone who cares more about HST students than Julie,” says Collin Stultz, the Nina T. and Robert H. Rubin Professor in Medical Engineering and Science, and professor of EECS. Stultz is also the co-director of HST, as well as a 1997 HST MD graduate. “She is, and has always been, a formidable advocate for HST students and an oracle of information to me.”
Elazer Edelman ’78, SM ’79, PhD ’84, the Edward J. Poitras Professor in Medical Engineering and Science and director of IMES, says that Greenberg “has been a mentor to generations of students and leaders — she is a force of nature whose passion for learning and teaching is matched by love for our people and the spirit of our institutions. Her name is synonymous with many of our most innovative educational initiatives; indeed, she has touched every aspect of HST and IMES this very many decades. It is hard to imagine academic life here without her guiding hand.”
Greenberg says she is looking forward to spending more time on her hobbies, including baking, gardening, and travel, and that she may investigate getting involved in some way with working with STEM and underserved communities. She describes leaving now as “bittersweet. But I think that HST is in a strong, secure position, and I’m excited to see what will happen next, but from further away … and as long as they keep inviting alumni to the HST dinners, I will come.”
#Alumni/ae#amazing#applications#Biological engineering#career#classes#communication#Community#computer#Computer Science#course#covid#development#education#Electrical Engineering&Computer Science (eecs)#Electronics#engineering#engineers#Faculty#Full#gardening#generations#Giving#Government#hand#Harvard-MIT Health Sciences and Technology#Health#Health sciences and technology#hearing#History
0 notes
Text
Researchers 3D print key components for a point-of-care mass spectrometer
New Post has been published on https://thedigitalinsider.com/researchers-3d-print-key-components-for-a-point-of-care-mass-spectrometer/
Researchers 3D print key components for a point-of-care mass spectrometer
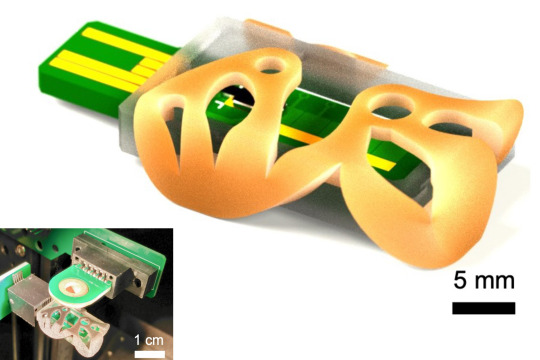

Mass spectrometry, a technique that can precisely identify the chemical components of a sample, could be used to monitor the health of people who suffer from chronic illnesses. For instance, a mass spectrometer can measure hormone levels in the blood of someone with hypothyroidism.
But mass spectrometers can cost several hundred thousand dollars, so these expensive machines are typically confined to laboratories where blood samples must be sent for testing. This inefficient process can make managing a chronic disease especially challenging.
“Our big vision is to make mass spectrometry local. For someone who has a chronic disease that requires constant monitoring, they could have something the size of a shoebox that they could use to do this test at home. For that to happen, the hardware has to be inexpensive,” says Luis Fernando Velásquez-García, a principal research scientist in MIT’s Microsystems Technology Laboratories (MTL).
He and his collaborators have taken a big step in that direction by 3D printing a low-cost ionizer — a critical component of all mass spectrometers — that performs twice as well as its state-of-the-art counterparts.
Their device, which is only a few centimeters in size, can be manufactured at scale in batches and then incorporated into a mass spectrometer using efficient, pick-and-place robotic assembly methods. Such mass production would make it cheaper than typical ionizers that often require manual labor, need expensive hardware to interface with the mass spectrometer, or must be built in a semiconductor clean room.
By 3D printing the device instead, the researchers were able to precisely control its shape and utilize special materials that helped boost its performance.
“This is a do-it-yourself approach to making an ionizer, but it is not a contraption held together with duct tape or a poor man’s version of the device. At the end of the day, it works better than devices made using expensive processes and specialized instruments, and anyone can be empowered to make it,” says Velásquez-García, senior author of a paper on the ionizer.
He wrote the paper with lead author Alex Kachkine, a mechanical engineering graduate student. The research is published in the Journal of the American Association for Mass Spectrometry.
Low-cost hardware
Mass spectrometers identify the contents of a sample by sorting charged particles, called ions, based on their mass-to-charge ratio. Since molecules in blood don’t have an electric charge, an ionizer is used to give them a charge before they are analyzed.
Most liquid ionizers do this using electrospray, which involves applying a high voltage to a liquid sample and then firing a thin jet of charged particles into the mass spectrometer. The more ionized particles in the spray, the more accurate the measurements will be.
The MIT researchers used 3D printing, along with some clever optimizations, to produce a low-cost electrospray emitter that outperformed state-of-the-art mass spectrometry ionizer versions.
They fabricated the emitter from metal using binder jetting, a 3D printing process in which a blanket of powdered material is showered with a polymer-based glue squirted through tiny nozzles to build an object layer by layer. The finished object is heated in an oven to evaporate the glue and then consolidate the object from a bed of powder that surrounds it.
“The process sounds complicated, but it is one of the original 3D printing methods, and it is highly precise and very effective,” Velásquez-García says.
Then, the printed emitters undergo an electropolishing step that sharpens it. Finally, each device is coated in zinc oxide nanowires which give the emitter a level of porosity that enables it to effectively filter and transport liquids.
Thinking outside the box
One possible problem that impacts electrospray emitters is the evaporation that can occur to the liquid sample during operation. The solvent might vaporize and clog the emitter, so engineers typically design emitters to limit evaporation.
Through modeling confirmed by experiments, the MIT team realized they could use evaporation to their advantage. They designed the emitters as externally-fed solid cones with a specific angle that leverages evaporation to strategically restrict the flow of liquid. In this way, the sample spray contains a higher ratio of charge-carrying molecules.
“We saw that evaporation can actually be a design knob that can help you optimize the performance,” he says.
They also rethought the counter-electrode that applies voltage to the sample. The team optimized its size and shape, using the same binder jetting method, so the electrode prevents arcing. Arcing, which occurs when electrical current jumps a gap between two electrodes, can damage electrodes or cause overheating.
Because their electrode is not prone to arcing, they can safely increase the applied voltage, which results in more ionized molecules and better performance.
They also created a low-cost, printed circuit board with built-in digital microfluidics, which the emitter is soldered to. The addition of digital microfluidics enables the ionizer to efficiently transport droplets of liquid.
Taken together, these optimizations enabled an electrospray emitter that could operate at a voltage 24 percent higher than state-of-the-art versions. This higher voltage enabled their device to more than double the signal-to-noise ratio.
In addition, their batch processing technique could be implemented at scale, which would significantly lower the cost of each emitter and go a long way toward making a point-of-care mass spectrometer an affordable reality.
“Going back to Guttenberg, once people had the ability to print their own things, the world changed completely. In a sense, this could be more of the same. We can give people the power to create the hardware they need in their daily lives,” he says.
Moving forward, the team wants to create a prototype that combines their ionizer with a 3D-printed mass filter they previously developed. The ionizer and mass filter are the key components of the device. They are also working to perfect 3D-printed vacuum pumps, which remain a major hurdle to printing an entire compact mass spectrometer.
“Miniaturization through advanced technology is slowly but surely transforming mass spectrometry, reducing manufacturing cost and increasing the range of applications. This work on fabricating electrospray sources by 3D printing also enhances signal strength, increasing sensitivity and signal-to-noise ratio and potentially opening the way to more widespread use in clinical diagnosis,” says Richard Syms, professor of microsystems technology in the Department of Electrical and Electronic Engineering at Imperial College London, who was not involved with this research.
This work was supported by Empiriko Corporation.
#3-D printing#3d#3D printing#additive manufacturing#applications#approach#Art#blood#board#box#chemical#chronic disease#college#Design#devices#diagnostics#direction#Disease#double#droplets#Electrical Engineering&Computer Science (eecs)#electrode#electrodes#electronic#Electronics#electrospray#engineering#engineers#filter#gap
0 notes
Text
A new computational technique could make it easier to engineer useful proteins
New Post has been published on https://thedigitalinsider.com/a-new-computational-technique-could-make-it-easier-to-engineer-useful-proteins/
A new computational technique could make it easier to engineer useful proteins


To engineer proteins with useful functions, researchers usually begin with a natural protein that has a desirable function, such as emitting fluorescent light, and put it through many rounds of random mutation that eventually generate an optimized version of the protein.
This process has yielded optimized versions of many important proteins, including green fluorescent protein (GFP). However, for other proteins, it has proven difficult to generate an optimized version. MIT researchers have now developed a computational approach that makes it easier to predict mutations that will lead to better proteins, based on a relatively small amount of data.
Using this model, the researchers generated proteins with mutations that were predicted to lead to improved versions of GFP and a protein from adeno-associated virus (AAV), which is used to deliver DNA for gene therapy. They hope it could also be used to develop additional tools for neuroscience research and medical applications.
“Protein design is a hard problem because the mapping from DNA sequence to protein structure and function is really complex. There might be a great protein 10 changes away in the sequence, but each intermediate change might correspond to a totally nonfunctional protein. It’s like trying to find your way to the river basin in a mountain range, when there are craggy peaks along the way that block your view. The current work tries to make the riverbed easier to find,” says Ila Fiete, a professor of brain and cognitive sciences at MIT, a member of MIT’s McGovern Institute for Brain Research, director of the K. Lisa Yang Integrative Computational Neuroscience Center, and one of the senior authors of the study.
Regina Barzilay, the School of Engineering Distinguished Professor for AI and Health at MIT, and Tommi Jaakkola, the Thomas Siebel Professor of Electrical Engineering and Computer Science at MIT, are also senior authors of an open-access paper on the work, which will be presented at the International Conference on Learning Representations in May. MIT graduate students Andrew Kirjner and Jason Yim are the lead authors of the study. Other authors include Shahar Bracha, an MIT postdoc, and Raman Samusevich, a graduate student at Czech Technical University.
Optimizing proteins
Many naturally occurring proteins have functions that could make them useful for research or medical applications, but they need a little extra engineering to optimize them. In this study, the researchers were originally interested in developing proteins that could be used in living cells as voltage indicators. These proteins, produced by some bacteria and algae, emit fluorescent light when an electric potential is detected. If engineered for use in mammalian cells, such proteins could allow researchers to measure neuron activity without using electrodes.
While decades of research have gone into engineering these proteins to produce a stronger fluorescent signal, on a faster timescale, they haven’t become effective enough for widespread use. Bracha, who works in Edward Boyden’s lab at the McGovern Institute, reached out to Fiete’s lab to see if they could work together on a computational approach that might help speed up the process of optimizing the proteins.
“This work exemplifies the human serendipity that characterizes so much science discovery,” Fiete says. “It grew out of the Yang Tan Collective retreat, a scientific meeting of researchers from multiple centers at MIT with distinct missions unified by the shared support of K. Lisa Yang. We learned that some of our interests and tools in modeling how brains learn and optimize could be applied in the totally different domain of protein design, as being practiced in the Boyden lab.”
For any given protein that researchers might want to optimize, there is a nearly infinite number of possible sequences that could generated by swapping in different amino acids at each point within the sequence. With so many possible variants, it is impossible to test all of them experimentally, so researchers have turned to computational modeling to try to predict which ones will work best.
In this study, the researchers set out to overcome those challenges, using data from GFP to develop and test a computational model that could predict better versions of the protein.
They began by training a type of model known as a convolutional neural network (CNN) on experimental data consisting of GFP sequences and their brightness — the feature that they wanted to optimize.
The model was able to create a “fitness landscape” — a three-dimensional map that depicts the fitness of a given protein and how much it differs from the original sequence — based on a relatively small amount of experimental data (from about 1,000 variants of GFP).
These landscapes contain peaks that represent fitter proteins and valleys that represent less fit proteins. Predicting the path that a protein needs to follow to reach the peaks of fitness can be difficult, because often a protein will need to undergo a mutation that makes it less fit before it reaches a nearby peak of higher fitness. To overcome this problem, the researchers used an existing computational technique to “smooth” the fitness landscape.
Once these small bumps in the landscape were smoothed, the researchers retrained the CNN model and found that it was able to reach greater fitness peaks more easily. The model was able to predict optimized GFP sequences that had as many as seven different amino acids from the protein sequence they started with, and the best of these proteins were estimated to be about 2.5 times fitter than the original.
“Once we have this landscape that represents what the model thinks is nearby, we smooth it out and then we retrain the model on the smoother version of the landscape,” Kirjner says. “Now there is a smooth path from your starting point to the top, which the model is now able to reach by iteratively making small improvements. The same is often impossible for unsmoothed landscapes.”
Proof-of-concept
The researchers also showed that this approach worked well in identifying new sequences for the viral capsid of adeno-associated virus (AAV), a viral vector that is commonly used to deliver DNA. In that case, they optimized the capsid for its ability to package a DNA payload.
“We used GFP and AAV as a proof-of-concept to show that this is a method that works on data sets that are very well-characterized, and because of that, it should be applicable to other protein engineering problems,” Bracha says.
The researchers now plan to use this computational technique on data that Bracha has been generating on voltage indicator proteins.
“Dozens of labs having been working on that for two decades, and still there isn’t anything better,” she says. “The hope is that now with generation of a smaller data set, we could train a model in silico and make predictions that could be better than the past two decades of manual testing.”
The research was funded, in part, by the U.S. National Science Foundation, the Machine Learning for Pharmaceutical Discovery and Synthesis consortium, the Abdul Latif Jameel Clinic for Machine Learning in Health, the DTRA Discovery of Medical Countermeasures Against New and Emerging threats program, the DARPA Accelerated Molecular Discovery program, the Sanofi Computational Antibody Design grant, the U.S. Office of Naval Research, the Howard Hughes Medical Institute, the National Institutes of Health, the K. Lisa Yang ICoN Center, and the K. Lisa Yang and Hock E. Tan Center for Molecular Therapeutics at MIT.
#000#acids#ai#algae#amino acids#applications#approach#Artificial Intelligence#Bacteria#Best Of#Biological engineering#Brain#Brain and cognitive sciences#brain research#brains#Cells#change#CNN#Collective#computer#Computer Science#Computer Science and Artificial Intelligence Laboratory (CSAIL)#Computer science and technology#conference#czech#data#Defense Advanced Research Projects Agency (DARPA)#Design#DNA#Electrical Engineering&Computer Science (eecs)
0 notes
Text
Researchers create “The Consensus Game” to elevate AI’s text comprehension and generation skills
New Post has been published on https://thedigitalinsider.com/researchers-create-the-consensus-game-to-elevate-ais-text-comprehension-and-generation-skills/
Researchers create “The Consensus Game” to elevate AI’s text comprehension and generation skills


Imagine you and a friend are playing a game where your goal is to communicate secret messages to each other using only cryptic sentences. Your friend’s job is to guess the secret message behind your sentences. Sometimes, you give clues directly, and other times, your friend has to guess the message by asking yes-or-no questions about the clues you’ve given. The challenge is, both of you want to make sure you’re understanding each other correctly and agreeing on the secret message.
MIT Computer Science and Artificial Intelligence Laboratory (CSAIL) researchers have created a similar “game” to help improve how AI understands and generates text. The “Consensus Game” involves two parts of an AI system — one part tries to generate sentences (like giving clues), and the other part tries to understand and evaluate those sentences (like guessing the secret message).
The researchers discovered that by treating this interaction as a game, where both parts of the AI work together under specific rules to agree on the right message, they could significantly improve the AI’s ability to give correct and coherent answers to questions. They tested this new game-like approach on a variety of tasks, such as reading comprehension, solving math problems, and carrying on conversations, and found that it helped the AI perform better across the board.
Traditionally, language models (LMs) answer one of two ways: generating answers directly from the model (generative querying) or using the model to score a set of predefined answers (discriminative querying), which can lead to differing and sometimes incompatible results. With the generative approach, “Who is the President of the United States?” might yield a straightforward answer like “Joe Biden.” However, a discriminative query could incorrectly dispute this fact when evaluating the same answer, such as “Barack Obama.”
So, how do we reconcile mutually incompatible scoring procedures to achieve coherent, efficient predictions?
“Imagine a new way to help language models understand and generate text, like a game. We’ve developed a training-free, game-theoretic method that treats the whole process as a complex game of clues and signals, where a generator tries to send the right message to a discriminator using natural language. Instead of chess pieces, they’re using words and sentences,” says MIT CSAIL PhD student Athul Jacob. “Our way to navigate this game is finding the ‘approximate equilibria,’ leading to a new decoding algorithm called ‘Equilibrium Ranking.’ It’s a pretty exciting demonstration of how bringing game-theoretic strategies into the mix can tackle some big challenges in making language models more reliable and consistent.”
When tested across many tasks, like reading comprehension, commonsense reasoning, math problem-solving, and dialogue, the team’s algorithm consistently improved how well these models performed. Using the ER algorithm with the LLaMA-7B model even outshone the results from much larger models. “Given that they are already competitive, that people have been working on it for a while, but the level of improvements we saw being able to outperform a model that’s 10 times the size was a pleasant surprise,” says Jacob.
Game on
Diplomacy, a strategic board game set in pre-World War I Europe, where players negotiate alliances, betray friends, and conquer territories without the use of dice — relying purely on skill, strategy, and interpersonal manipulation — recently had a second coming. In November 2022, computer scientists, including Jacob, developed “Cicero,” an AI agent that achieves human-level capabilities in the mixed-motive seven-player game, which requires the same aforementioned skills, but with natural language. The math behind this partially inspired The Consensus Game.
While the history of AI agents long predates when OpenAI’s software entered the chat (and never looked back) in November 2022, it’s well documented that they can still cosplay as your well-meaning, yet pathological friend.
The Consensus Game system reaches equilibrium as an agreement, ensuring accuracy and fidelity to the model’s original insights. To achieve this, the method iteratively adjusts the interactions between the generative and discriminative components until they reach a consensus on an answer that accurately reflects reality and aligns with their initial beliefs. This approach effectively bridges the gap between the two querying methods.
In practice, implementing the Consensus Game approach to language model querying, especially for question-answering tasks, does involve significant computational challenges. For example, when using datasets like MMLU, which have thousands of questions and multiple-choice answers, the model must apply the mechanism to each query. Then, it must reach a consensus between the generative and discriminative components for every question and its possible answers.
The system did struggle with a grade school right of passage: math word problems. It couldn’t generate wrong answers, which is a critical component of understanding the process of coming up with the right one.
“The last few years have seen really impressive progress in both strategic decision-making and language generation from AI systems, but we’re just starting to figure out how to put the two together. Equilibrium ranking is a first step in this direction, but I think there’s a lot we’ll be able to do to scale this up to more complex problems.”
An avenue of future work involves enhancing the base model by integrating the outputs of the current method. This is particularly promising since it can yield more factual and consistent answers across various tasks, including factuality and open-ended generation. The potential for such a method to significantly improve the base model’s performance is high, which could result in more reliable and factual outputs from ChatGPT and similar language models that people use daily.
“Even though modern language models, such as ChatGPT and Gemini, have led to solving various tasks through chat interfaces, the statistical decoding process that generates a response from such models has remained unchanged for decades,” says Google research scientist Ahmad Beirami. “The proposal by the MIT researchers is an innovative game-theoretic framework for decoding from language models through solving the equilibrium of a consensus game. The significant performance gains reported in the research paper are promising, opening the door to a potential paradigm shift in language model decoding that may fuel a flurry of new applications.”
Jacob wrote the paper with MIT-IBM Watson Lab researcher Yikang Shen and MIT Department of Electrical Engineering and Computer Science assistant professors Gabriele Farina and Jacob Andreas, who is also a CSAIL member. They will present their work at the International Conference on Learning Representations (ICLR) this May. The research received a “best paper award” at the NeurIPS R0-FoMo Workshop in December and it will also be highlighted as a “spotlight paper” at ICLR.
#2022#agreement#ai#ai agent#AI systems#algorithm#Algorithms#applications#approach#artificial#Artificial Intelligence#biden#board#challenge#chatGPT#chess#comprehension#computer#Computer Science#Computer Science and Artificial Intelligence Laboratory (CSAIL)#Computer science and technology#conference#datasets#december#direction#Electrical Engineering&Computer Science (eecs)#engineering#Europe#framework#fuel
0 notes
Text
Second round of seed grants awarded to MIT scholars studying the impact and applications of generative AI
New Post has been published on https://thedigitalinsider.com/second-round-of-seed-grants-awarded-to-mit-scholars-studying-the-impact-and-applications-of-generative-ai/
Second round of seed grants awarded to MIT scholars studying the impact and applications of generative AI


Last summer, MIT President Sally Kornbluth and Provost Cynthia Barnhart issued a call for papers to “articulate effective roadmaps, policy recommendations, and calls for action across the broad domain of generative AI.” The response to the call far exceeded expectations with 75 proposals submitted. Of those, 27 proposals were selected for seed funding.
In light of this enthusiastic response, Kornbluth and Barnhart announced a second call for proposals this fall.
“The groundswell of interest and the caliber of the ideas overall made clear that a second round was in order,” they said in their email to MIT’s research community this fall. This second call for proposals resulted in 53 submissions.
Following the second call, the faculty committee from the first round considered the proposals and selected 16 proposals to receive exploratory funding. Co-authored by interdisciplinary teams of faculty and researchers affiliated with all five of the Institute’s schools and the MIT Schwarzman College of Computing, the proposals offer insights and perspectives on the potential impact and applications of generative AI across a broad range of topics and disciplines.
Each selected research group will receive between $50,000 and $70,000 to create 10-page impact papers. Those papers will be shared widely via a publication venue managed and hosted by the MIT Press under the auspices of the MIT Open Publishing Services program.
As with the first round of papers, Thomas Tull, a member of the MIT School of Engineering Dean’s Advisory Council and a former innovation scholar at the School of Engineering, contributed funding to support the effort.
The selected papers are:
“A Road-map for End-to-end Privacy and Verifiability in Generative AI,” led by Alex Pentland, Srini Devadas, Lalana Kagal, and Vinod Vaikuntanathan;
“A Virtuous Cycle: Generative AI and Discovery in the Physical Sciences,” led by Philip Harris and Phiala Shanahan;
“Artificial Cambrian Intelligence: Generating New Forms of Visual Intelligence,” led by Ramesh Raskar and Tomaso A. Poggio;
“Artificial Fictions and the Value of AI-Generated Art,” led by Justin Khoo;
“GenAI for Improving Human-to-human Interactions with a Focus on Negotiations,” led by Lawrence Susskind;
“Generative AI as a New Applications Platform and Ecosystem,” led by Michael Cusumano;
“Generative AI for Cities: A Civic Engagement Playbook,” led by Sarah Williams, Sara Beery, and Eden Medina;
“Generative AI for Textile Engineering: Advanced Materials from Heritage Lace Craft,” led by Svetlana V. Boriskina;
“Generative AI Impact for Biomedical Innovation and Drug Discovery,” led by Manolis Kellis, Brad Pentelute, and Marinka Zitnik;
“Impact of Generative AI on the Creative Economy,” led by Ashia Wilson and Dylan Hadfield-Menell;
“Redefining Virtuosity: The Role of Generative AI in Live Music Performances,” led by Joseph A. Paradiso and Eran Egozy;
“Reflection-based Learning with Generative AI,” led by Stefanie Mueller;
“Robust and Reliable Systems for Generative AI,” led by Shafi Goldwasser, Yael Kalai, and Vinod Vaikuntanathan;
“Supporting the Aging Population with Generative AI,” led by Pattie Maes;
“The Science of Language in the Era of Generative AI,” led by Danny Fox, Yoon Kim, and Roger Levy; and
“Visual Artists, Technological Shock, and Generative AI,” led by Caroline Jones and Huma Gupta.
#000#Administration#advanced materials#aging#ai#Algorithms#applications#Art#artificial#Artificial Intelligence#artists#cities#college#Community#Computer science and technology#computing#craft#drug#drug discovery#economy#Electrical Engineering&Computer Science (eecs)#email#engineering#Faculty#focus#Forms#Funding#genai#generative#generative ai
0 notes