#drug discovery
Link
Scientists at St. Jude Children’s Research Hospital have published their findings on SJ3149, a chemical with broad efficacy against various cancer types, particularly acute myeloid leukemia (AML). Scientists worked together to discover and improve a chemical molecule that functions as a molecular “super-glue” for a variety of tumors. SJ3149 binds to cancer-related protein casein kinase 1 alpha (CK1α), causing its elimination. The findings were published in the journal Nature Communications.
Have you ever wished you could just put a “Do Not Enter” sign on a faulty protein in a cancer cell? Thanks to molecular glue degraders, scientists are coming closer to doing just that. These ingenious tiny molecules function as matchmakers, bringing together a protein designated for destruction (the target) and the cellular executioner (an E3 ligase) in a lethal embrace. As a result, the target protein is tagged and disposed of, providing a fresh approach to cancer treatment.
Molecular glues work by hijacking the cell’s natural protein recycling system. The molecular glue binds the targeted protein to an enzyme, marking it for destruction via proteasomal degradation. For many cancer-related proteins that conventional small molecule inhibitors cannot effectively target, molecular glues may be a feasible therapeutic option. This prompted scientists at St. Jude to create a vast proprietary library of molecular glues and screen it against a variety of cancer cell lines, resulting in an early hit.
Continue Reading
138 notes
·
View notes
Text
Using single-cell genomics to accelerate drug development for pulmonary illnesses

- By InnoNurse Staff -
A group of researchers used single-cell genomics, which captures gene activities in individual cells, to examine the activity of all cells in hPCLS after various experimental and therapeutic treatments.
Read more at Helmholtz Association of German Research Centres/Medical Xpress
2 notes
·
View notes
Photo

Professor Anne Osbourn has been awarded the 2023 Novozymes Prize for her pioneering work in helping to produce important drugs in greater volumes and improving the natural defence systems of plants.
Her discovery that plant genes for specialised pathways are organised like beads on a string has fuelled the finding of novel plant compounds and pathways.
Professor Graham Moore Director of the John Innes Centre says: “I am delighted that the innovative work and distinguished career of Anne Osbourn has been recognised with this prestigious award. It reflects Anne’s huge contribution to the exciting field of natural product biosynthesis and in providing solutions for human medicine and plant health.”
(Source: The John Innes Centre Linked In page)
https://www.jic.ac.uk/press-release/professor-anne-osbourn-plant-scientist-and-poet-receives-prestigious-novozymes-prize/?utm_campaign=Oktopost-NEWS+-+Prof+Anne+Osbourn+-+Novozymes&utm_content=Oktopost-linkedin&utm_medium=social&utm_source=linkedin
#women in science#scientist#katia plant scientist#botanist#plant biology#genetics#molecular biology#biosynthesis#orchid#artist#drug discovery#plant science#biology#women in stem
10 notes
·
View notes
Text
Epigenomic analysis sheds light on risk factors for ALS
New Post has been published on https://thedigitalinsider.com/epigenomic-analysis-sheds-light-on-risk-factors-for-als/
Epigenomic analysis sheds light on risk factors for ALS

For most patients, it’s unknown exactly what causes amyotrophic lateral sclerosis (ALS), a disease characterized by degeneration of motor neurons that impairs muscle control and eventually leads to death.
Studies have identified certain genes that confer a higher risk of the disease, but scientists believe there are many more genetic risk factors that have yet to be discovered. One reason why these drivers have been hard to find is that some are found in very few patients, making it hard to pick them out without a very large sample of patients. Additionally, some of the risk may be driven by epigenomic factors, rather than mutations in protein-coding genes.
Working with the Answer ALS consortium, a team of MIT researchers has analyzed epigenetic modifications — tags that determine which genes are turned on in a cell — in motor neurons derived from induced pluripotent stem (IPS) cells from 380 ALS patients.
This analysis revealed a strong differential signal associated with a known subtype of ALS, and about 30 locations with modifications that appear to be linked to rates of disease progression in ALS patients. The findings may help scientists develop new treatments that are targeted to patients with certain genetic risk factors.
“If the root causes are different for all these different versions of the disease, the drugs will be very different and the signals in IPS cells will be very different,” says Ernest Fraenkel, the Grover M. Hermann Professor in Health Sciences and Technology in MIT’s Department of Biological Engineering and the senior author of the study. “We may get to a point in a decade or so where we don’t even think of ALS as one disease, where there are drugs that are treating specific types of ALS that only work for one group of patients and not for another.”
MIT postdoc Stanislav Tsitkov is the lead author of the paper, which appears today in Nature Communications.
Finding risk factors
ALS is a rare disease that is estimated to affect about 30,000 people in the United States. One of the challenges in studying the disease is that while genetic variants are believed to account for about 50 percent of ALS risk (with environmental factors making up the rest), most of the variants that contribute to that risk have not been identified.
Similar to Alzheimer’s disease, there may be a large number of genetic variants that can confer risk, but each individual patient may carry only a small number of those. This makes it difficult to identify the risk factors unless scientists have a very large population of patients to analyze.
“Because we expect the disease to be heterogeneous, you need to have large numbers of patients before you can pick up on signals like this. To really be able to classify the subtypes of disease, we’re going to need to look at a lot of people,” Fraenkel says.
About 10 years ago, the Answer ALS consortium began to collect large numbers of patient samples, which could allow for larger-scale studies that might reveal some of the genetic drivers of the disease. From blood samples, researchers can create induced pluripotent stem cells and then induce them to differentiate into motor neurons, the cells most affected by ALS.
“We don’t think all ALS patients are going to be the same, just like all cancers are not the same. And the goal is being able to find drivers of the disease that could be therapeutic targets,” Fraenkel says.
In this study, Fraenkel and his colleagues wanted to see if patient-derived cells could offer any information about molecular differences that are relevant to ALS. They focused on epigenomic modifications, using a method called ATAC-seq to measure chromatin density across the genome of each cell. Chromatin is a complex of DNA and proteins that determines which genes are accessible to be transcribed by the cell, depending on how densely packed the chromatin is.
In data that were collected and analyzed over several years, the researchers did not find any global signal that clearly differentiated the 380 ALS patients in their study from 80 healthy control subjects. However, they did find a strong differential signal associated with a subtype of ALS, characterized by a genetic mutation in the C9orf72 gene.
Additionally, they identified about 30 regions that were associated with slower rates of disease progression in ALS patients. Many of these regions are located near genes related to the cellular inflammatory response; interestingly, several of the identified genes have also been implicated in other neurodegenerative diseases, such as Parkinson’s disease.
“You can use a small number of these epigenomic regions and look at the intensity of the signal there, and predict how quickly someone’s disease will progress. That really validates the hypothesis that the epigenomics can be used as a filter to better understand the contribution of the person’s genome,” Fraenkel says.
“By harnessing the very large number of participant samples and extensive data collected by the Answer ALS Consortium, these studies were able to rigorously test whether the observed changes might be artifacts related to the techniques of sample collection, storage, processing, and analysis, or truly reflective of important biology,” says Lyle Ostrow, an associate professor of neurology at the Lewis Katz School of Medicine at Temple University, who was not involved in the study. “They developed standard ways to control for these variables, to make sure the results can be accurately compared. Such studies are incredibly important for accelerating ALS therapy development, as they will enable data and samples collected from different studies to be analyzed together.”
Targeted drugs
The researchers now hope to further investigate these genomic regions and see how they might drive different aspects of ALS progression in different subsets of patients. This could help scientists develop drugs that might work in different groups of patients, and help them identify which patients should be chosen for clinical trials of those drugs, based on genetic or epigenetic markers.
Last year, the U.S. Food and Drug Administration approved a drug called tofersen, which can be used in ALS patients with a mutation in a gene called SOD1. This drug is very effective for those patients, who make up about 1 percent of the total population of people with ALS. Fraenkel’s hope is that more drugs can be developed for, and tested in, people with other genetic drivers of ALS.
“If you had a drug like tofersen that works for 1 percent of patients and you just gave it to a typical phase two clinical trial, you probably wouldn’t have anybody with that mutation in the trial, and it would’ve failed. And so that drug, which is a lifesaver for people, would never have gotten through,” Fraenkel says.
The MIT team is now using an approach called quantitative trait locus (QTL) analysis to try to identify subgroups of ALS patients whose disease is driven by specific genomic variants.
“We can integrate the genomics, the transcriptomics, and the epigenomics, as a way to find subgroups of ALS patients who have distinct phenotypic signatures from other ALS patients and healthy controls,” Tsitkov says. “We have already found a few potential hits in that direction.”
The research was funded by the Answer ALS program, which is supported by the Robert Packard Center for ALS Research at Johns Hopkins University, Travelers Insurance, ALS Finding a Cure Foundation, Stay Strong Vs. ALS, Answer ALS Foundation, Microsoft, Caterpillar Foundation, American Airlines, Team Gleason, the U.S. National Institutes of Health, Fishman Family Foundation, Aviators Against ALS, AbbVie Foundation, Chan Zuckerberg Initiative, ALS Association, National Football League, F. Prime, M. Armstrong, Bruce Edwards Foundation, the Judith and Jean Pape Adams Charitable Foundation, Muscular Dystrophy Association, Les Turner ALS Foundation, PGA Tour, Gates Ventures, and Bari Lipp Foundation. This work was also supported, in part, by grants from the National Institutes of Health and the MIT-GSK Gertrude B. Elion Research Fellowship Program for Drug Discovery and Disease.
#000#Administration#als#American Airlines#amyotrophic lateral sclerosis#Amyotrophic lateral sclerosis (ALS)#Analysis#approach#Biological engineering#Biology#blood#caterpillar#cell#Cells#chemical#chromatin#coding#communications#data#development#direction#Disease#Diseases#DNA#drug#drug discovery#drugs#engineering#Environmental#filter
0 notes
Text
Unleashing the Power of Generative AI in Drug Discovery: A Game-Changer in Healthcare

In the realm of drug discovery, where innovation is key to saving lives, a new hero has emerged — Generative AI. With its transformative capabilities, Generative AI is revolutionizing the process of developing new medications, offering unprecedented speed and precision. Let’s delve into how this cutting-edge technology is reshaping the landscape of drug discovery in healthcare.
What is Generative AI?
Generative AI is like a creative wizard in the world of artificial intelligence. It has the magical ability to learn from existing data and generate new content based on this knowledge. In the case of drug discovery, Generative AI acts as a virtual laboratory assistant, swiftly proposing novel drug compounds and predicting their effectiveness.

How Generative AI Works in Drug Discovery?
Generative AI in drug discovery analyzes vast amounts of chemical data to create potential drug candidates. By understanding patterns and structures within this data, the AI can generate new compounds, optimize them for specific targets, and predict their efficacy. This process accelerates the identification of promising drug candidates and reduces the time and resources required for traditional trial-and-error methods.
Applications of Generative AI in Drug Discovery
Accelerating Drug Discovery: Generative AI significantly speeds up the drug discovery process by suggesting new compounds and predicting their properties, allowing researchers to focus on the most promising candidates.
Optimizing Drug Design: The technology helps in optimizing drug structures for enhanced potency and reduced side effects, leading to the development of safer and more effective medications.
Personalizing Treatment: Generative AI enables personalized medicine by tailoring drug therapies based on individual genetic profiles, offering customized treatment plans for better patient outcomes.
Generative AI has brought about numerous game-changing applications in healthcare, transforming processes, simplifying tasks, and reducing errors across the industry. Here, find out more about how generative AI is making a significant impact in the field of healthcare with its transformative capabilities.
Conclusion
Generative AI stands as a beacon of hope in the field of drug discovery, unlocking new possibilities and propelling healthcare into a future of enhanced precision and efficiency. By harnessing the power of AI-driven innovation, we are bridging the gap between science fiction and reality, bringing us closer to a world where life-saving medications are just a discovery away.
#generative ai#artificial intelligence#drug discovery#application of generative AI#healthcare#healthcare technology#technology#technologies
0 notes
Text
Africaʼs First Integrated Drug Discovery and Development Platform.

Africaʼs first integrated drug discovery and development center. H3D was founded at UCT in April 2010 and focuses on translational medicine, which involves early-stage medicine discovery in the lab through to the treatment of patients in clinical settings. WIPO Magazine recently sat down with Chibale to learn more about H3D, and the role intellectual property plays in its groundbreaking work.
WM: What is the potential of drug discovery in Africa?
KC: Africa is arguably the most genetically diverse continent.
Everybody came from Africa and went somewhere else. That means diseases are not African problems or African diseases, they are human diseases, human problems. So, drug discovery in Africa has huge potential to contribute to humanity and to create local jobs.
And how is H3D affecting health innovation in Africa?
H3D is having an impact at various levels, particularly by creating drug discovery infrastructure and platforms capable of contributing to the global pipeline of innovative products that could be further developed. In other words, we have strengthened our capacity to translate basic science knowledge into potential life-saving medicines. And we are bridging the gap between the lab and the patient.
You focused initially on malaria. Why?
Malaria was an opportunity for us to build the infrastructure required for translational medicine. At the end of the day, beyond understanding the biology of the human malaria parasite, the drug discovery principles are the same for malaria or cancer. For example, regardless of the disease, among other things, the common goal is to understand how the human body will handle the drug candidate. The malaria project was an opportunity to work with the Medicines for Malaria Venture (MMV) and to subsequently engage with new partners, such as Merck and the Bill and Melinda Gates Foundation. Once we developed the infrastructure we needed for that project, we began adding other diseases, including tuberculosis (TB), and antimicrobial resistance. In 2022, we had an opportunity to work with Johnson & Johnson as one of the companyʼs three satellite centers for global health discovery. In sum, malaria was an anchor program that enabled us to acquire the skills and experience we wanted to develop, and which we then transferred to other diseases.
How important are such partnerships to H3Dʼs work, and to
developing a robust health innovation ecosystem in Africa?
Partnerships are extremely important, even for innovative pharmaceutical companies with financial muscle. Indeed, some of the product portfolios they offer include drug candidates licensed in from third parties. This enables them to de-risk the early stages of drug development. For H3D, partnerships were important from the start, for three reasons. First, to tackle infrastructure challenges; second, to build the technology platforms we needed; and third, to access skilled people. Partnerships are also important to secure funding. When you have a project with global support, you attract partners who share the same goals, funding grows, and you gain access to a network of centers of excellence. Partnerships can bring to the table what you donʼt have, because everyone is interested in the projectʼs success. When there is mutual interest, you can make a huge difference.
What about the importance of building a local procurement
support system?
One of the main barriers to scientific innovation in Africa has been a lack of infrastructure in the broad sense. This includes a local procurement support system with functioning laboratories, access to the spare parts you need when something breaks down, the ability to access reagents and chemicals readily and rapidly, and so on.
Of course, from a business perspective, we need scale that justifies the business. At present, there are too few players, so business opportunities are limited. Thatʼs why weʼre working to expand the community to create the demand that will foster the businesses we need to supply the chemicals and reagents required for research and development, for example.

What is the role of intellectual property in all this?
When thereʼs an unmet medical need, you have to innovate, and IP incentivizes innovation. IP is an enabler and underpins robust innovation ecosystems. Cash-strapped universities can use IP to generate new sources of income from their research, through university spinouts, for example. IP is also a magnet for investment. People want to invest in a country where there is respect for rules and laws, including IP.
Do you still need IP in Africa for infectious diseases, where
commercial returns are low?
Absolutely. Because IP is also a responsibility, even for infectious diseases where commercial returns are perceived to be low. Without IP you would have a free-for-all. When it comes to health equity, itʼs important to remember that the person who owns the IP can decide whether to share it voluntarily or not.When you hold IP rights in a medicine, you can control its use to some extent. Thatʼs why, in Africa, we need to be owning IP. When we do, and we find an appropriate partner to take the IP forward, we get a return. I would rather own 1% of one billion than 99.99% of zero.
IP is also a responsibility, even for infectious diseases where commercial returns are perceived to be low.
What is the current focus of H3Dʼs work?
In terms of drug discovery, weʼre focusing on action studies to identify biological targets and to better understand the mechanism of resistance of these targeted organisms to drugs. These organisms are very clever. Our job is to outsmart them.
Do you still see the need for new approaches?
Yes. At a scientific level, Iʼm an advocate for Afro-centric drug discovery. You need to find a target to hit – an enzyme or a protein – which may respond differently in different populations for genetic reasons.

Genetic differences in the expression and activity of drug-metabolizing
enzymes can lead to variable responses to therapeutics. For example, in people of African descent, due to genetic mutations, enzymes responsible for metabolizing the antiretroviral drug Efavirenz work more slowly than in other populations and this can result in toxicity, even death, due to drug overdose if dosages arenʼt adjusted appropriately. So, drug development needs to move from a one-size-fits-all focus toward a population-centric approach.
We really need to invest in understanding the genetics of the African
population with respect to biological drug targets we go after and the
enzymes responsible for metabolizing specific drugs. Also, we need to address the funding gap in translational medicine, which
many investors find too risky. This will require policy changes to encourage investors to see drug development as a continuum that requires investment at each stage of the value chain. This would create opportunities to share both risks and benefits, and ultimately will benefit everyone.
Read the full interview online and learn more about Chibaleʼs recommendations for developing a robust health innovation system in Africa.

#medicine discovery#intellectual property#health sector#drug discovery#health innovation#26 april#worldipday#pharmaceutical#drug development#sdg3#ip and the sdgs
0 notes
Text
Biotech: Xaira Therapeutics raises $1 billion in funding

- By Nuadox Crew -
U.S.-based Xaira Therapeutics, a newly launched biotechnology company, aims to revolutionize the drug discovery and development process by leveraging advanced AI technologies.
Co-founded by ARCH Venture Partners, Foresite Labs, and Dr. David Baker of UW Medicine, Xaira launches with over $1 billion in funding.
Led by Dr. Marc Tessier-Lavigne, former Genentech CSO, the company integrates AI research, expansive data generation, and therapeutic product development to accelerate the delivery of effective therapies.
--
Source: Xaira Therapeutics
Read Also
This AI model directly compares the properties of prospective new pharmaceuticals
#medtech#biotech#drug discovery#medicine#Xaira Therapeutics#pharma#health tech#ai#artificial intelligence
0 notes
Text

MAGNA™: A Revolutionary New Technology for Studying Protein-Protein Interactions
#Protein-Protein Interactions (PPIs)#Drug Discovery#MAGNA™ Platform#Biological Research#Disease Research
0 notes
Text
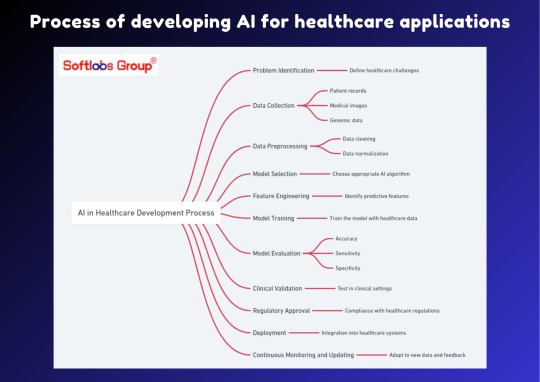
Discover the process of developing AI for healthcare applications with our informative guide. This simplified overview outlines the essential steps involved in leveraging artificial intelligence to enhance healthcare solutions. Perfect for anyone interested in the intersection of technology and healthcare. Stay informed with Softlabs Group for more insightful content on cutting-edge advancements in AI.
0 notes
Text
Behavioral Assays
Assessing neurological traits and events in rodents is an essential part of neurological research, and behavioral testing plays a vital role in evaluating these factors. These tests, which measure traits such as locomotor activity, depression-like behavior, socialization, memory, and many others, provide a complementary perspective to imaging studies in evaluating brain function. At Ekam, we offer a suite of behavioral tests to provide a comprehensive assessment of neurological function in animal models, alongside our imaging services.
0 notes
Link
In 2023, bioinformatics discoveries have catalyzed immense progress across the life sciences landscape. These ground-breaking discoveries have provided insight into the complex workings of biological systems, processes, and disease states. From discovering new diagnostic markers to mapping the complexity of the brain, these innovations promise to transform medicine, evolution, and beyond. The pace of bioinformatics discoveries fueled by the rise of artificial intelligence heralds a new era of opportunity. In this article, we take a closer look at 10 of the best bioinformatics innovations of the year and their profound impact on the field of biology. Get ready for an exciting journey to the greatest discoveries in bioinformatics!
#1 CRACKING THE CODE OF MYSTERIOUS “Y” CHROMOSOME
Scientists have struggled for decades to sequence the enigmatic Y chromosome essential to male biology. The Telomere-to-Telomere (T2T) consortium presented the complete sequence of the human Y chromosome, encompassing 62,460,029 base pairs from the HG002 genome (T2T-Y). Thanks to new computational techniques, they can finally peer into its genetic blueprint and understand male infertility and human evolution like never before.
Continue Reading
57 notes
·
View notes
Text
AI drug discovery company Xaira emerges with $1 billion in funding

- By InnoNurse Staff -
Industry heavyweights including ex-FDA Commissioner Scott Gottlieb, M.D., Nobel Prize winner Carolyn Bertozzi, Ph.D., and former Johnson & Johnson CEO Alex Gorsky, are among the prominent figures on the board of Xaira Therapeutics.
Read more at Fierce Biotech
///
Other recent news and insights
New initiative launched to support health-tech innovators in Africa (Disrupt Africa)
Artificial intelligence capable of assessing cardiovascular risk during CT scans (Cedars Sinai)
#ai#drug discovery#xaira therapeutics#medtech#biotech#health tech#startups#africa#innovation#ct scan#radiology#cardiology#diagnostics#imaging
0 notes
Text
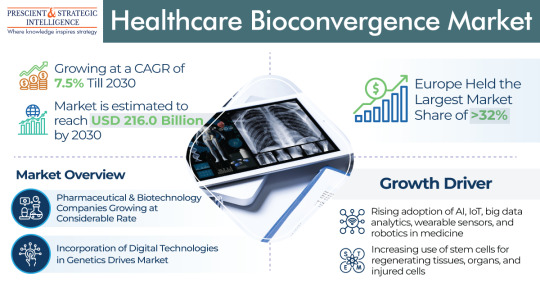
Drug Discovery was the Leading Category in the Healthcare Bioconvergence Industry
The healthcare bioconvergence market will power at a rate of 7.5% rate by the end of this decade, to touch USD 216 billion by 2030.The industry is growing due to the growing count of elderly people and the growing usage of stem cells for regenerating organs, tissues, and injured cells.
Moreover, the preference of users for combinations of cutting-edge technologies, including ML, ergonomics,…
View On WordPress
#Bioelectronic#Diagnostic and Biological Sensors#Drug Discovery#Healthcare Bioconvergence#Nanorobotics for Drug Delivery#Regenerative Medicine
0 notes
Text
Serotonina e psichedelici: sono allucinogeni solo se si accoppiano al segnale cellulare giusto
L’interesse per le sostanze psichedeliche classiche o serotoninergiche è riemerso, dato il loro potenziale di indurre effetti terapeutici rapidi e prolungati. Le sostanze psichedeliche sono limitate dai loro effetti allucinogeni e possono causare confusione e ansia. Sebbene un recente studio preclinico abbia suggerito la possibilità di separare le sostanze psichedeliche dalle loro proprietà…

View On WordPress
#allucinogeno#cellular signaling#drug discovery#farmaco#farmacoforo#G protein#psichedelico#recettore#Serotonina#triptammina
0 notes
Text
New Developments to Support Global Growth of the Drug Discovery Services Industry
This Study Will Help You:# To define, characterize, and project the Drug Discovery Services Market according to end user, drug type, process, and geography.
# To give comprehensive details regarding the main elements (drivers, constraints, opportunities, and challenges) affecting the market’s growth.
# To strategically examine micromarkets in light of their respective contributions to the…
View On WordPress
#Drug Discovery#Drug Discovery Services#Drug Discovery Services Industry#Drug Discovery Services Market#Global Drug Discovery Services#Global Drug Discovery Services Industry#Global Drug Discovery Services Market
0 notes