#replication terminator protein
Text
After growing crystals of highly purified RTP combined with a short stretch of duplex DNA, the X-ray diffraction pattern can be used to obtain a detailed picture of the atoms involved in the RTP/DNA complex (figure 25.14).
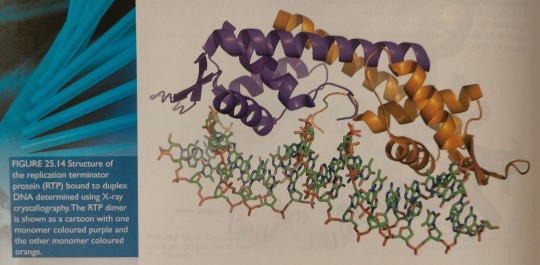
"Chemistry" 2e - Blackman, A., Bottle, S., Schmid, S., Mocerino, M., Wille, U.
#book quotes#chemistry#nonfiction#textbook#crystals#rtp#replication terminator protein#bacillus subtilis#x ray crystallography#duplex dna#dna#deoxyribonucleic acid#atoms#chemical bonding
0 notes
Text
It is a dimeric protein that binds with exceptionally high affinity to duplex DNA at specific nucleotide sequences forming a termination complex that prevents further DNA replication.
"Chemistry" 2e - Blackman, A., Bottle, S., Schmid, S., Mocerino, M., Wille, U.
#book quote#chemistry#nonfiction#textbook#replication terminator protein#rtp#bacillus subtilis#dimeric#protein#chemical bonding#dna#deoxyribonucleic acid#nucleotide#dna replication#termination
0 notes
Text

Immense pride, tinged with sadness.
For those who would like to read the full list:
1908 MECHNIKOV, ELIE
FOR THEIR WORK ON IMMUNITY
1908 EHRLICH, PAUL
FOR THEIR WORK ON IMMUNITY
1914 BARANY, ROBERT
FOR HIS WORK ON THE PHYSIOLOGY AND PATHOLOGY OF THE VESTIBULAR APPARATUS
1922 MEYERHOF, OTTO FRITZ
FOR HIS DISCOVERY OF THE FIXED RELATIONSHIP BETWEEN THE CONSUMPTION OF
OXYGEN AND THE METABOLISM OF LACTIC ACID IN THE MUSCLE
1930 LANDSTEINER, KARL
FOR HIS DISCOVERY OF HUMAN BLOOD GROUPS
1936 LOEWI, OTTO
FOR THEIR DISCOVERIES RELATING TO CHEMICAL TRANSMISSION OF NERVE IMPULSES
1944 ERLANGER, JOSEPH
FOR THEIR DISCOVERIES RELATING TO THE HIGHLY DIFFERENTIATED FUNCTIONS OF SINGLE NERVE FIBRES
1945 CHAIN, ERNST BORIS
FOR THE DISCOVERY OF PENICILLIN AND ITS CURATIVE EFFECT IN VARIOUS INFECTIOUS DISEASES
1946 MULLER, HERMANN J.
FOR THE DISCOVERY OF THE PRODUCTION OF MUTATIONS BY MEANS OF X-RAY IRRADIATION
1947 CORI, GERTY THERESA, RADNITZ
FOR THEIR DISCOVERY OF THE COURSE OF THE CATALYTIC CONVERSION OF GLYCOGEN
1950 REICHSTEIN, TADEUS
FOR THEIR DISCOVERIES RELATING TO THE HORMONES OF THE ADRENAL CORTEX, THEIR STRUCTURE AND BIOLOGICAL EFFECTS
1952 WAKSMAN, SELMAN A.
FOR HIS DISCOVERY OF STREPTOMYCIN, THE FIRST ANTIBIOTIC EFFECTIVE AGAINST TUBERCULOSIS
1953 LIPMANN, FRITZ ALBERT
FOR HIS DISCOVERY OF CO-ENZYME A AND ITS IMPORTANCE FOR INTERMEDIARY METABOLISM
1953 KREBS, HANS ADOLF
FOR HIS DISCOVERY OF THE CITRIC ACID CYCLE
1958 LEDERBERG, JOSHUA
FOR HIS DISCOVERIES CONCERNING GENETIC RECOMBINATION AND THE ORGANISATION OF THE GENETIC MATERIAL OF BACTERIA
1959 KORNBERG, ARTHUR
FOR THEIR DISCOVERY OF THE MECHANISMS IN THE BIOLOGICAL SYNTHESIS OF RIBONUCLEIC ACID AND DEOXYRIBONUCLEIC ACID
1964 BLOCH, KONRAD
FOR THEIR DISCOVERIES CONCERNING THE MECHANISM AND REGULATION OF THE CHOLESTEROL AND FATTY ACID METABOLISM
1965 JACOB, FRANCOIS
FOR THEIR DISCOVERIES CONCERNING GENETIC CONTROL OF ENZYME AND VIRUS SYNTHESIS
1965 LWOFF, ANDRE
FOR THEIR DISCOVERIES CONCERNING GENETIC CONTROL OF ENZYME AND VIRUS SYNTHESIS
1967 WALD, GEORGE
FOR THEIR DISCOVERIES CONCERNING THE PRIMARY PHYSIOLOGICAL AND CHEMICAL VISUAL PROCESSES IN THE EYE
1968 NIRENBERG, MARSHALL W.
FOR THEIR INTERPRETATION OF THE GENETIC CODE AND ITS FUNCTION IN PROTEIN SYNTHESIS
1969 LURIA, SALVADOR E.
FOR THEIR DISCOVERIES CONCERNING THE REPLICATION MECHANISM AND THE GENETIC STRUCTURE OF VIRUSES
1970 KATZ, BERNARD
FOR THEIR DISCOVERIES CONCERNING THE HUMORAL TRANSMITTERS IN THE NERVE TERMINALS AND THE MECHANISM
FOR THEIR STORAGE, RELEASE AND INACTIVATION
1970 AXELROD, JULIUS
FOR THEIR DISCOVERIES CONCERNING THE HUMORAL TRANSMITTERS IN THE NERVE TERMINALS AND THE MECHANISM
FOR THEIR STORAGE, RELEASE AND INACTIVATION
1972 EDELMAN, GERALD M.
FOR THEIR DISCOVERIES CONCERNING THE CHEMICAL STRUCTURE OF ANTIBODIES
1975 TEMIN, HOWARD M.
FOR THEIR DISCOVERIES CONCERNING THE INTERACTION BETWEEN TUMOR VIRUSES AND THE GENETIC MATERIAL OF THE CELL
1975 BALTIMORE, DAVID
FOR THEIR DISCOVERIES CONCERNING THE INTERACTION BETWEEN TUMOR VIRUSES AND THE GENETIC MATERIAL OF THE CELL
1976 BLUMBERG, BARUCH S.
FOR THEIR DISCOVERIES CONCERNING NEW MECHANISMS FOR THE ORIGIN AND DISSEMINATION OF INFECTIOUS DISEASES
1977 YALOW, ROSALYN
FOR THE DEVELOPMENT OF RADIOIMMUNOASSAYS OF PEPTIDE HORMONES
1977 SCHALLY, ANDREW V.
FOR THEIR DISCOVERIES CONCERNING THE PEPTIDE HORMONE PRODUCTION OF THE BRAIN
1978 NATHANS, DANIEL
FOR THE DISCOVERY OF RESTRICTION ENZYMES AND THEIR APPLICATION TO PROBLEMS OF MOLECULAR GENETICS
1980 BENACERRAF, BARUJ
FOR THEIR DISCOVERIES CONCERNING GENETICALLY DETERMINED STRUCTURES ON THE CELL SURFACE THAT
REGULATE IMMUNOLOGICAL REACTIONS
1984 MILSTEIN, CESAR
FOR THEORIES CONCERNING THE SPECIFICITY IN DEVELOPMENT AND CONTROL OF THE IMMUNE SYSTEM AND THE DISCOVERY OF THE
PRINCIPLE FOR PRODUCTION OF MONOCLONAL ANTIBODIES
1985 BROWN, MICHAEL S.
FOR THEIR DISCOVERIES CONCERNING THE REGULATION OF CHOLESTEROL METABOLISM
1985 GOLDSTEIN, JOSEPH L.
FOR THEIR DISCOVERIES CONCERNING THE REGULATION OF CHOLESTEROL METABOLISM
1986 COHEN, STANLEY
FOR THEIR DISCOVERIES OF GROWTH FACTORS
1986 LEVI-MONTALCINI, RITA
FOR THEIR DISCOVERIES OF GROWTH FACTORS
1988 ELION, GERTRUDE B.
FOR THEIR DISCOVERIES OF IMPORTANT PRINCIPLES FOR DRUG TREATMENT
1989 VARMUS, HAROLD E.
FOR THEIR DISCOVERY OF THE CELLULAR ORIGIN OF RETROVIRAL ONCOGENES
1994 RODBELL, MARTIN
FOR THEIR DISCOVERY OF G-PROTEINS AND THE ROLE OF THESE PROTEINS IN SIGNAL TRANSDUCTION IN CELLS
1994 GILMAN, ALFRED G.
FOR THEIR DISCOVERY OF G-PROTEINS AND THE ROLE OF THESE PROTEINS IN SIGNAL TRANSDUCTION IN CELLS
1997 PRUSINER, STANLEY B.
FOR HIS DISCOVERY OF PRIONS - A NEW BIOLOGICAL PRINCIPLE OF INFECTION
1998 FURCHGOTT, ROBERT F.
FOR THEIR DISCOVERIES CONCERNING NITRIC OXIDE AS A SIGNALING MOLECULE IN THE CARDIOVASCULAR SYSTEM
2000 GREENGARD, PAUL
FOR THEIR DISCOVERIES CONCERNING SIGNAL TRANSDUCTION IN THE NERVOUS SYSTEM
2000 KANDEL, ERIC R.
FOR THEIR DISCOVERIES CONCERNING SIGNAL TRANSDUCTION IN THE NERVOUS SYSTEM
2002 BRENNER, SYDNEY
FOR THEIR DISCOVERIES CONCERNING GENETIC REGULATION OF ORGAN DEVELOPMENT AND PROGRAMMED CELL DEATH
2002 HORVITZ, H. ROBERT
FOR THEIR DISCOVERIES CONCERNING GENETIC REGULATION OF ORGAN DEVELOPMENT AND PROGRAMMED CELL DEATH
2004 AXEL, RICHARD
FOR THEIR DISCOVERIES OF ODORANT RECEPTORS AND THE ORGANIZATION OF THE OLFACTORY SYSTEM
2006 FIRE, ANDREW Z.
FOR THEIR DISCOVERY OF RNA INTERFERENCE - GENE SILENCING BY DOUBLE-STRANDED RNA
2011 STEINMAN, RALPH M.
FOR THEIR DISCOVERIES CONCERNING THE ACTIVATION OF INNATE IMMUNITY
2011 BEUTLER, BRUCE A.
FOR THEIR DISCOVERIES CONCERNING THE ACTIVATION OF INNATE IMMUNITY
2013 SCHEKMAN, RANDY W.
FOR THEIR DISCOVERIES OF MACHINERY REGULATING VESICLE TRAFFIC, A MAJOR TRANSPORT SYSTEM IN OUR CELLS
2013 ROTHMAN, JAMES E.
FOR THEIR DISCOVERIES OF MACHINERY REGULATING VESICLE TRAFFIC, A MAJOR TRANSPORT SYSTEM IN OUR CELLS
2017 ROSBASH, MICHAEL
FOR THEIR DISCOVERIES OF MOLECULAR MECHANISMS CONTROLLING THE CIRCADIAN RHYTHM
Likud Herut UK
81 notes
·
View notes
Text
Histone Modifications
Hello, hello! Today's topic is histone modifications. We are continuing on with the epigenetics theme after my previous educational post about DNA methylation. As described in that post, epigenetics is the study of heritable genetic modifications without a change in DNA sequence (Takuno & Gaut, 2012). Similarly to DNA methylation, histone modifications affect gene expression through regulation of accessibility of the DNA for transcription (Bartova et al, 2008). But before we get into these modifications, let's go over a bit of background information!
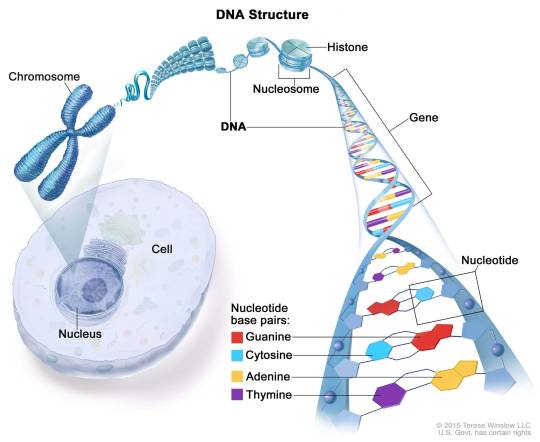
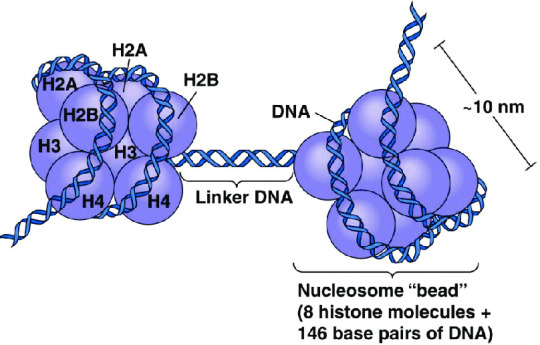
What is a histone, anyway? A histone is a type of protein involved in DNA compaction and organization. In order to fit a genome's worth of DNA into the nucleus of a cell, that stuff needs to be extremely tightly packed! Histones help with this by forming an octomer called a nucleosome, which the DNA wraps around. These nucleosomes then coil together to form a fiber known as chromatin, which goes on to make up a chromosome. When the chromatin is less tightly packed, it is known as euchromatin and it is available for transcription (Bartova et al, 2008). When it is more tightly packed, it is known as heterchromatin, and polymerase proteins cannot access and transcribe the DNA (Bartova et al, 2008). Histone modifications regulate the transition between heterochromatin and euchromatin (Bartova et al, 2008).
(Above image from humanoriginproject.com)
(Above image from Caputi et al, 2017)
The octomer core of a nucleosome is made up of two copies of each of four types of histones: H2A, H2B, H3, and H4 (Marino-Ramirez et al, 2017). Each of these histones includes an N-terminal tail structure, which is the main site of modification (Marino-Ramirez et al, 2017). The tails are modified through addition and removal of certain functional groups or other small structures. Types of modifications include acetylation by histone acetyltransferases, methylation by histone methyltransferases, phosphorylation by kinases, and ubiquitination (Marino-Ramirez et al, 2017). All of this information is used for naming specific histone modifications: Which histone is modified, which amino acid of the histone tail the modification is on, what type of modification is made, and in what amount. For example, H3K9me2 is the name for di-methylation of the 9th Lysine on an H3 histone's tail.
Some important histone modifications and their effects include:
H3K9me2: transcriptional activation + maintenance of CHG DNA methylation in plants
H3K9me3: transcriptional repression
H3K9ac: transcriptional activation
H3K4me1 & H3K4me3: transcriptional activation
H3K27me3: transcriptional repression
H4K16ac: transcriptional activation
H3S10p: DNA replication-related chromatin condensation
(He & Lehming, 2003)
Important Terms: histone, nucleosome, heterochromatin, euchromatin, transcription, epigenetics
4 notes
·
View notes
Text
Terminal...
The S phase was the next step in the interphase. This was when the DNA replicated itself, ensuring that each new cell that would be formed would have a complete and identical copy of the original. As the DNA doubled in length, the cell paid close attention to the process, making sure that each strand was perfectly aligned with its counterpart. If any errors were detected, specialized proteins called DNA polymerases would attempt to repair the damage. If the damage was too extensive, the cell would initiate a process called apoptosis, ensuring that the damaged DNA did not pass on to future generations.
2 notes
·
View notes
Text
Lasso peptides are a really cool family of peptides that have a very unique knotted structure and are chronically understudied and poorly understood. Lasso peptides get their name from the way the N-terminus wraps back around the peptide chain and forms an isopeptide bond with the carbonyl of a Glu/Asp residue, forming a literal lasso like shape:

This is a deceptively tricky reaction to study, let alone replicate. At the minute, most studies done on identifying and isolating these molecules use overexpression, chemical or even biosynthetic strategies are inaccessible, which is a real shame because the unusual topologies of these peptides gives them excellent protease resistance and much better pharmacokinetics than linear, or even standard cyclic peptides.
More detailed images of the peptides reveal some interesting quirks of how they adopt this unique shape:

This stick model on the left show the whole lasso peptide, with the section forming the ring highlighted in green. The sequence that is ‘lasso’d’ so to speak is quite interesting, and is shown on the right. The C terminal tryptophan-phenylalanine motif is common among lasso peptides and acts as a ‘stopper’ and provides a, frankly massive, steric barrier to unfolding the lasso. The encapsulated section is comprised of glycine which is pretty easy to rationalise because glycine doesn’t have a side chain, thus making it the most suitable for threading through a ring. I’ve also highlighted the proline, which is also common among lasso peptides, it is though that the proline is important to preorganising the lasso before the ring is completed, proline is a much more restricted amino acid than the others and often adopts this role as a conformational guide.
Its worth remembering just how tight this whole arrangement is, threading a peptide though a 7 residue ring is no small feat, and here’s the space filling model to highlight that fact:

There is no wiggle room at all, and that is key to understanding why these things are so hard to make, they incur a huge entropic and steric penalties for being in these tightly wound conformations and aren’t offsetting that by forming covalent bonds or massive protein scale stabilising interactions. As such I think they’re pretty remarkable and definitely worth studying more.
References:
1. Tan, S., Moore, G., and Nodwell, J. (2019) Put a bow on it: Knotted antibiotics take center stage. Antibiotics 8, 117.
2. Liu, T., Ma, X., Yu, J., Yang, W., Wang, G., Wang, Z., Ge, Y., Song, J., Han, H., Zhang, W., Yang, D., Liu, X., and Ma, M. (2021) Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications. Chemical Science 12, 12353–12364.
3. Structure, bioactivity, and resistance mechanism of streptomonomicin, an unusual lasso peptide from an understudied halophilic actinomycete. Chemistry & Biology. Cell Press.
4 notes
·
View notes
Text
"the name of the beast"
parasitic reaction in the hindbrain
long-forgotten viruses, microglial cascades,
cells singing with love for something that
never lived. Hyperflat meets misfolded
protein, self-replicating prions infest
the cerebellum, the heart, the small
and large intestine. Everything is a
metaphor. Everything in the world
is exactly the same. Diseases gentle
and warm take us in tender embraces
and usher us to a state of rest. The
growing pains of globalization settle
easy in the soft bones of children,
hard in joints made firm by age
We can put it all behind us:
I am thinking, and you are thinking with me,
and it could be faster, we could be closer,
If you'd only find a way to let your guard
down, let the light in. Repentance, they say,
is as good as medicine. Cheaper, besides.
(febrile from the poison some hired
goon sprayed into my brain, careless,
like hot glue used to set a bone, maybe
nothing but hot glue used to set a bone
fractured by a burrsaw for the intromission
of wires. Like a bedbug mating, spraying
the stuff of life into a living creature with
a needle and cauter so like a penis. We know
human beings. Human beings love wires and
they don't care who suffers or how, who
dies or how. One creature's good as another
If I could do it all over again, if I could
do it all over again, if I could do it all
over again, darling, I'd never have let
them take me alive)
I am thinking, and the machine is thinking
with me. The machine is thinking, and I am
thinking with it. I am thinking, and the machine
is thinking with me. I am thinking, and the
machine is thinking with me
the children first, whose minds nature
cast in some unlovable contour. The sick
next, who we must learn to see as a burden
to make peace. When we meet the men who
remake us, it will be face to face, with eyes
like ours, but cold and born to command;
it will be with one mind, one heart,
one seat of thought and spirit. They
will untangle the tongues God once confused
into the straight and perfect english of
the terminal, and our obedience to
our masters' voices will be before
thought. Faster, anyway, than prayer,
with thoroughly documented behaviors
and perfectly anticipated responses
a safe environment in which we can
be trusted, at last, and for all time
12 notes
·
View notes
Text
LOOK AT IT READ IT AND NOODLE ON IT
(NSHL) Arab-Israeli family bearing 12S rRNA (m.1555A>G) mutation (Bykhovskaya et al., 2004). The m.1555A>G mutation, locates in the decoding site of the mitochondrial small subunit (SSU) ribosomal RNA, is the first identified homoplasmic mitochondrial mutation. In addition, the mutation is predicted to cause an alteration in the second structure, which impairs protein synthesis and enlarges sensitivity to aminoglycoside ototoxicity (Prezant et al., 1993). It is also well accepted that m.1555A>G mutation presents as a key cause of antibiotic-induced hearing loss; however, the mutation alone typically does not lead to disease. Among these Arab-Israeli family members without previous exposure to aminoglycosides, the m.1555A>G mutation induced various clinical phenotypes ranging from severe congenital deafness, to moderate progressive hearing loss of later onset, to completely normal hearing. It was characterized by Guan et al. that there’s more severe biochemical defects in the lymphoblastoid cells derived from symptomatic individuals than those from asymptomatic ones of the Arab-Israeli family bearing m.1555A>G mutation (Guan et al., 1996). On the other hand, an identical degree of mitochondrial dysfunction was observed when they compared the cybrids cell lines derived from symptomatic and asymptomatic individuals (Guan et al., 2001). These findings strongly indicate that the m.A1555G mutation as a primary cause of hearing loss and nuclear modifier genes play a role in modulating the phenotypic expression (Guan et al., 2006). Naturally, the most promising candidates would be these nuclear genes encode the subunits of respiratory chain complex, proteins involved in mitochondrial protein synthesis, and proteins involved in mtDNA replication and maintenance. TFB1M (transcription factor B1), encoding a mitochondrial rRNA methyltransferase, has been putatively identified as a possible nuclear modifier of the m.1555A>G mutation, suggesting a connection between 12S rRNA methylation and hearing loss (Raimundo et al., 2012).
Mutations in mitochondrial tRNAs have been reported to be associated with various mitochondrial disease states (Abbott et al., 2014). With disrupted structures, mt tRNAs mutations would cause defective translation and impaired mt protein synthesis, leading to defects in OXPHOS systems. Post-transcriptional processing, including maturation of primary tRNA, multiple chemical residue modifications, and aminoacylation, are critical to accurate and effective translation. Thus enzymes involved in these processing are highly possible modifier genes. The penetrance is much higher in the presence of nuclear mutations involved in transfer RNA base modification (MTO1, TRMU-MTO2, and GTPBP3 genes) (Guan et al., 2006, Li and Guan, 2003, Li et al., 2002); however, additional supporting evidence is still needed to firmly confirm their role as genetic modifier. Establishment of ideal animal models may help discover their functions in mitochondrial diseases and explain their tissue specificity. Recent studies largely expand the phenotypic spectrum associated with different aminoacyl-tRNA synthetases (ARS). McMilan et al. reported congenital visual impairment and progressive microcephaly has been associated with KARS mutations (McMillan et al., 2014) and Nakajima et al. reported a homozygous YARS2 causes severe myopathy, lactic acidosis, and sideroblastic anemia 2 (Nakajima et al., 2014). Another whole-exome sequencing study reveals that mutations in VASR2 and TARS2 are the causes of mitochondrial encephalomyopathies (Diodato et al., 2014). Perli et al. further reported that isolated non-catalytic C-terminal of LASR2 can improve both viability and bioenergetic proficiency of cybrid cells carrying pathogenic mutations in mt-tRNAs (Perli et al., 2014). These findings strongly suggest the group of aminoacyl-tRNA synthetases as active modifying players in mitochondrial disorders, and may lead to further understanding of tissue specific mitochondrial diseases.
In general, our knowledge of modifier genes involved in mitochondrial disorders has increased substantially in the past decade. Several mitochondrial rRNA methyltransferase and mitochondrial tRNA modifications have been identified in human, but the proteins involved in these modifications are far from being all identified. Understanding how the cells modulate biological processes to accommodate the adverse effects of mtDNA dysfunction is important as it may provide vital clues in the search for modifier genes as well as therapeutic targets.

2 notes
·
View notes
Text
Fwd: Graduate position: UOttawa.Three.MolEvolution
Begin forwarded message:
> From: [email protected]
> Subject: Graduate position: UOttawa.Three.MolEvolution
> Date: 3 April 2024 at 05:55:39 BST
> To: [email protected]
>
>
> Dear All,
>
> I have three PhD positions in my lab. My research areas are roughly
> split into
>
> 1) Molecular mechanisms in translation/transcription and RNA
> processing/genome replication through comparative studies, focusing on
> how molecular machines carry out their functions by decoding signals,
> e.g., release factors decoding termination signals, spliceosomes decoding
> splicing signals, etc., and how these signals and decoders coevolve.
>
> 2) Microbiology and infectious diseases, with a focus on host-parasite
> coevolution at the molecular level.
>
> 3) Molecular evolution/bioinformatics, i.e., how proteins, DNA, and RNA
> interact with each other and how the interaction partners change over
> time, leading to functional changes and adaptation at the molecular level.
>
> 4) Molecular phylogenetics and phylogeography to understand the
> distribution of life over space and time, with a focus on invasive
> species and how they evolve in the new environment.
>
> You might first browse the recent articles from my laboratory at:
> https://ift.tt/Qq3k8KN
>
> You can then let me know which article is the closest to your interest. We
> can schedule a zoom meeting to discuss the details.
>
> Best
> Xuhua Xia
> University of Ottawa, Canada
> https://ift.tt/53IC6XJ
> https://ift.tt/b2fqHKR
> https://ift.tt/UqfPVwA
>
>
> Xuhua Xia
0 notes
Text
Post-Catalytic Complexes with Emtricitabine or Stavudine and HIV-1 Reverse Transcriptase Reveal New Mechanistic Insights for Nucleotide Incorporation and Drug Resistance
Since its discovery in the early 1980s, the human immunodeficiency virus 1 (HIV-1) has been a major health issue with nearly 38.0 million people infected globally in 2019 according to the WHO [1]. Despite extensive research efforts, neither a cure nor a vaccine for HIV-1 infection has been discovered yet. However, a breakthrough has been achieved with the highly active antiretroviral therapy (HAART), which significantly improves the life expectancy for patients with acquired immune deficiency syndrome (AIDS).
Two of the six classes of the United States Food and Drug Administration (FDA) approved drugs for HIV-1 treatment target the reverse transcriptase (RT) protein, an enzyme critical for the replication cycle of HIV-1 [2]. These two classes are nucleoside RT inhibitors (NRTIs) and non-nucleoside RT inhibitors (NNRTIs). While NNRTIs are allosteric inhibitors that alter the chemical catalysis rate limiting step through conformational changes [3,4], NRTIs mimic nucleotides that bind to the active site of RT. Since NRTIs lack the 30 -hydroxyl group necessary for chain elongation, their incorporation results in termination of the viral DNA transcription [5]. NRTIs are essential components of HAART and part of almost all FDA approved combination therapies for the treatment and protection of an infection with HIV. However, some FDA-approved NRTIs are now rarely prescribed (e.g., stavudine (d4T)) or discontinued (e.g., zalcitabine (ddC)) due to their off-target toxicity.
0 notes
Text
📆 10 Nov 2021 📰 Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2
Individuals with potential exposure to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) do not necessarily develop PCR or antibody positivity, suggesting that some individuals may clear subclinical infection before seroconversion. T cells can contribute to the rapid clearance of SARS-CoV-2 and other coronavirus infections1,2,3. Here we hypothesize that pre-existing memory T cell responses, with cross-protective potential against SARS-CoV-2 (refs. 4,5,6,7,8,9,10,11), would expand in vivo to support rapid viral control, aborting infection.
We measured SARS-CoV-2-reactive T cells, including those against the early transcribed replication–transcription complex (RTC)12,13, in intensively monitored healthcare workers (HCWs) who tested repeatedly negative according to PCR, antibody binding and neutralization assays (seronegative HCWs (SN-HCWs)). SN-HCWs had stronger, more multispecific memory T cells compared with a cohort of unexposed individuals from before the pandemic (prepandemic cohort), and these cells were more frequently directed against the RTC than the structural-protein-dominated responses observed after detectable infection (matched concurrent cohort).
SN-HCWs with the strongest RTC-specific T cells had an increase in IFI27, a robust early innate signature of SARS-CoV-2 (ref. 14), suggesting abortive infection. RNA polymerase within RTC was the largest region of high sequence conservation across human seasonal coronaviruses (HCoV) and SARS-CoV-2 clades. RNA polymerase was preferentially targeted (among the regions tested) by T cells from prepandemic cohorts and SN-HCWs. RTC-epitope-specific T cells that cross-recognized HCoV variants were identified in SN-HCWs. Enriched pre-existing RNA-polymerase-specific T cells expanded in vivo to preferentially accumulate in the memory response after putative abortive compared to overt SARS-CoV-2 infection. Our data highlight RTC-specific T cells as targets for vaccines against endemic and emerging Coronaviridae.

There is wide variability in the outcome of exposure to SARS-CoV-2, ranging from severe illness to asymptomatic infection, to those individuals who remain negative according to standard diagnostic tests. Recent studies have identified SARS-CoV-2 T cell reactivity in prepandemic samples5,6,7,8,9,10,11,15,16,17,18 and isolated cases of exposed individuals who have not seroconverted with single-time-point screening4,16,19,20,21,22. We studied an intensively monitored cohort of HCWs with potential exposure during the first UK pandemic wave (23 March 2020), comparing those with or without PCR and/or antibody evidence of SARS-CoV-2 infection.
We postulated that, in HCWs for whom PCR and the most sensitive binding and neutralizing antibody tests remained repeatedly negative (SN-HCWs), T cell assays might distinguish a subset of SN-HCWs with a subclinical, rapidly terminated (abortive) infection. We hypothesized that these individuals would exhibit pre-existing memory T cells with cross-reactive potential, obviating the time required for de novo T cell priming and clonal expansion.
In SN-HCWs, and in an additionally recruited cohort of medical students and laboratory staff with stored prepandemic samples that remained seronegative after close contact with cases, we had the opportunity to compare SARS-CoV-2-specific memory T cells with those that were already present in the same individual before, or at the time of, potential exposure.
We included an analysis of the understudied T cells directed against the core RTC within open reading frame 1ab (ORF1ab) (RNA polymerase co-factor non-structural protein 7 (NSP7), RNA polymerase NSP12 and helicase NSP13, hereafter the RTC); these are putative targets for pre-existing responses with pan-Coronaviridae reactivity, because they are likely to be highly conserved due to their key early roles in the viral life cycle.
Consistent with this, in cases in which immunity against other viruses (including hepatitis B virus (HBV), hepatitis C virus (HCV), HIV and Japaneses encephalitis virus (JEV)) has been described in exposed seronegative individuals, T cells were more likely to target non-structural proteins, such as polymerase, compared with in individuals with a seropositive infection23,24,25,26,27.
0 notes
Text
13th August in the Medical History

I. Introduction
Born on August 13, 1918, Frederick Sanger emerged as an English chemist whose indelible contributions profoundly impacted medical history. His ground-breaking work and novel approaches transformed our understanding of biological structures and made a lasting impact on the field. Sanger’s exceptional scientific journey encompassed two Nobel Prizes, earned for his elucidation of insulin’s structure in 1958 and his pioneering DNA sequencing techniques in 1980. We discover a visionary scientist whose legacy continues to influence the field of medical research and diagnostic improvements as we look deeper into Sanger’s life and accomplishments.
II. Early Life and Education

Frederick Sanger’s formative years, born into a modest family on August 13, 1918, cultivated his scientific curiosity. His early education at Bryanston School and subsequent studies at His outstanding career was founded at St. John’s College in Cambridge. At the University of Cambridge’s Biochemistry Department, Sanger’s aptitude for chemistry and strict work ethic were apparent. He developed his abilities and laid the groundwork for his significant contributions to medical science under the tutelage of eminent experts. In addition to developing his intellectual prowess, these early experiences gave him the dedication and tenacity that would characterize his ground-breaking research activities.
III. Achievements and Contributions in the Workplace

The career of Frederick Sanger was marked by pivotal discoveries that had an impact on the course of medicine. His groundbreaking study of insulin’s composition, which earned him the 1958 Nobel Prize, shed important light on how hormones work and might cause diabetes. Sanger won a second Nobel Prize in 1980 for his brilliant DNA sequencing technology, which had the greatest impact of all. This technique fundamentally changed genetics by making it possible to decode the complex code of DNA. Its extensive applications ranged from advancing the Human Genome Project to comprehending genetic diseases. A cornerstone in the structure of medical research and innovation, Sanger’s unflinching dedication to scientific excellence left an everlasting imprint.
A. First Nobel Prize: Insulin Structure (1958)
1. Explanation of Sanger’s role in determining the structure of insulin
Sanger’s diligent use of protein sequencing methods revealed the precise arrangement of amino acids in insulin, illuminating its intricate structure. He revolutionized the management of diabetes and advanced our understanding of hormone biology by deducing the order of amino acids in the chains of insulin. Sanger’s revolutionary work earned him the 1958 Nobel Prize in Chemistry, highlighting his significant contribution to the development of medical science.
2. Effect of this discovery on managing and understanding diabetes
Sanger’s discovery of the structure of insulin served as the foundation for the creation of more efficient diabetes treatments. Understanding the structure of insulin made it easier to create synthetic analogs with more therapeutic potential. This discovery was a turning point in the treatment of diabetes, resulting in new insulin formulations that better control blood sugar levels. Additionally, knowing more about the structure of insulin helped us better grasp how hormones and diabetes interact, advancing medical research and treatment methods.
B. DNA Sequencing Innovations
1. Introduction to Sanger sequencing method
Frederick Sanger’s ground-breaking method of Sanger sequencing, which allowed for the precise reading of DNA sequences, revolutionized genetic science. This technique, which is based on DNA replication and termination, enables the precise determination of nucleotide sequences. The addition of fluorescent colors increased its effectiveness even more. Sanger sequencing has had a significant impact on genetics, diagnosis, and evolutionary biology, influencing how we investigate and comprehend the genetic code of life.
2.Detailed explanation of the history and importance of the approach
Frederick Sanger’s DNA sequencing method, established in the 1970s, hinged on DNA replication and chain termination. He produced a number of fragmented DNA strands that were connected by chain-terminating dideoxynucleotides and matched various sequence locations. Gel electrophoresis then sorted these fragments by size, revealing the DNA sequence. The Human Genome Project and other significant scientific achievements, like the deciphering of DNA codes, were made possible by this ground-breaking technique, which changed genetics research.
3.DNA sequencing applications in medical research and diagnostics
The influence of DNA sequencing on medical research and diagnostics is significant. It has transformed our understanding of genetic disease by making it easier to pinpoint the mutations that cause disorders. Sequencing is used by cancer genomics to direct treatment choices. Genetic malformations are uncovered via prenatal testing, which influences reproductive decisions. Infectious disease outbreaks are tracked through pathogen genome sequencing. DNA sequencing has become a pillar of contemporary medical practice thanks to these uses, which have enhanced personalized medicine, early illness detection, and precision therapeutics.
C. Second Nobel Prize: DNA Sequencing (1980)

1. Recognition of Sanger’s achievements in DNA sequencing
Frederick Sanger’s contributions to DNA sequencing represent a pivotal turning point in the development of science. His method’s accuracy and dependability altered how we can decode genetic data, opening up new directions in genetics study and medical practice. The Nobel Prize was awarded for this discovery, honoring his crucial contribution to fundamentally altering how we understand genetics. A genomic revolution was sparked by Sanger’s sequencing method, enabling innumerable academics and practitioners to explore the intricate details of life’s genetic code.
2. Impact of his work on genetics research and the Human Genome Project
The Human Genome Project and genetics research have been forever changed as a result of Frederick Sanger’s efforts. His technique for DNA sequencing laid the groundwork for the project’s pivotal large-scale genome sequencing. Sanger’s method greatly expedited the exploration of the genome by enabling the quick and accurate decoding of DNA. This essential contribution not only accelerated the Human Genome Project but also opened the door for further genetic advancements that shaped medical research and customized medicine.
IV. Legacy and Influence

The influence of Frederick Sanger permeates numerous scientific fields. His ground-breaking DNA sequencing technique sparked revolutionary advances in genomics, advancing everything from medical diagnostics to evolutionary studies. The Human Genome Project was made possible by Sanger’s work, which significantly changed our understanding of genetics and disease. His precise research methodology, commitment to precision, and innovative attitude have inspired scientists all across the world. His legacy is upheld through the Sanger Institute, which promotes genetic and genomic research. Sanger’s enormous contributions to understanding the complexity of life’s genetic code are still felt in the genomic era, which is a monument to his lasting influence.
V. Role in Medical History

Frederick Sanger had a pioneering role in the history of medicine, whose scientific discoveries transformed how we understand biology and genetics. His ground-breaking discovery on the structure of insulin paved the path for more effective diabetes therapies and advanced hormone studies. A genomic revolution with applications in illness diagnosis, treatment, and customized medicine was sparked by his brilliant DNA sequencing approach, which also ushered genetics study into the current era. Sanger’s influence cuts across academic boundaries, influencing medical research by giving scientists the means to decipher the genetic code of life and unravel the riddles of health and illness.
VI. Conclusion

Finally, it should be noted that Frederick Sanger made unequalled contributions to the history of medicine. His discoveries on the structure of insulin and DNA sequencing fundamentally changed how we think about and manage illnesses, ushering in a time of individualized care and cutting-edge study. Sanger left a lasting legacy that has motivated subsequent generations of scientists to solve the genetic puzzle. Sanger’s contributions are further demonstrated by innovations like the QMe Hospital Management and Information System (HMIS), which streamlines patient care, improves data management, and enhances medical services while preserving Sanger’s legacy of pushing the boundaries of medical science.
0 notes
Text
Bacillus subtilis encodes a discrete flap endonuclease that cleaves #RNA-DNA hybrids
by Frances Caroline Lowder, Lyle A. Simmons
The current model for Okazaki fragment maturation in bacteria invokes #RNA cleavage by RNase H, followed by strand displacement synthesis and 5′ #RNA flap removal by DNA polymerase I (Pol I). #RNA removal by Pol I is thought to occur through the 5′-3′ flap endo/exonuclease (FEN) domain, located in the N-terminus of the protein. In addition to Pol I, many bacteria encode a second, Pol I-independent FEN. The contribution of Pol I and Pol I-independent FENs to DNA replication and genome stability remains unclear. In this work we purified Bacillus subtilis Pol I and FEN, then assayed these proteins on a variety of #RNA-DNA hybrid and DNA-only substrates. We found that FEN is far more active than Pol I on nicked double-flap, 5′ single flap, and nicked #RNA-DNA hybrid substrates. We show that the 5′ nuclease activity of B. subtilis Pol I is feeble, even during DNA synthesis when a 5′ flapped substrate is formed modeling an Okazaki fragment intermediate. Examination of Pol I and FEN on DNA-only substrates shows that FEN is more active than Pol I on most substrates tested. Further experiments show that ΔpolA phenotypes are completely rescued by expressing the C-terminal polymerase domain while expression of the N-terminal 5′ nuclease domain fails to complement ΔpolA. Cells lacking FEN (ΔfenA) show a phenotype in conjunction with an RNase HIII defect, providing genetic evidence for the involvement of FEN in Okazaki fragment processing. With these results, we propose a model where cells remove #RNA primers using FEN while upstream Okazaki fragments are extended through synthesis by Pol I. Our model resembles Okazaki fragment processing in eukaryotes, where Pol δ catalyzes strand displacement synthesis followed by 5′ flap cleavage using FEN-1. Together our work highlights the conservation of ordered steps for Okazaki fragment processing in cells ranging from bacteria to human. https://journals.plos.org/plosgenetics/article?id=10.1371%2Fjournal.pgen.1010585&utm_source=dlvr.it&utm_medium=tumblr
0 notes
Text
Two Faces of Adenovirus - A Time to Kill, A Time to Heal
Viruses are a kind of non-cellular organisms that are tiny to the nanometer level, with a simple structure that only has a protein shell and a long nucleic acid chain as genetic material. It cannot be independently metabolized but must be parasitic in living cells and multiply in a replication mode. It is weak but also strong, and human beings have to face the two-sided nature of a kind.
Adenovirus is such a virus that can infect human body. They are widespread in nature and can cause infections of the respiratory tract, eyes, gastrointestinal tract, and urinary system. They can cause regional transmission in highly confined, crowded and humid environments. But the advancement of science and technology helps us discover that this type of virus can also be used to fight tumors and even the novel coronavirus disease, COVID-19, that has recently raged around the world.
· Adenovirus Infection Can Be Dangerous
Adenovirus infection will cause many uncomfortable symptoms, including symptoms of upper respiratory tract infection (fever, sore throat, nasal congestion and other common cold symptoms), conjunctivitis (such as pharyngeal conjunctivitis, conjunctivitis, epidemic keratoconjunctivitis), gastrointestinal infection (diarrhea, more common in children under 5 years old), urinary tract infections, hemorrhagic cystitis, etc.
Adenovirus infection is a contagious disease and should be actively prevented. Adenovirus transmission mainly includes contact transmissions of respiratory droplets, pollutants and excrement, and eye secretions.
· Adenovirus Help Fight Tumors
Oncolytic viruses are currently an important method in the field of tumor immunotherapy. At present, Adenoviruses are the most used type in clinical trials. Oncolytic adenovirus is a genetically engineered adenovirus that can selectively replicate and express in tumor cells, thereby lysing tumor cells.
In 1996, since the world's first oncolytic adenovirus ONXY-015 was launched in clinical research, it has been widely used in scientific research, involving a variety of solid tumors. Oncolytic adenovirus combined with radiotherapy, chemotherapy, and targeted drug therapy are also proved to be more effective treatment options. Further clinical research will continue to explore the potential of oncolytic virus combined with other therapies.
· Adenovirus Vaccine for SARS-CoV-2
Adenovirus-vectored COVID-19 vaccines are one of the hot topics in global news. Johnson & Johnson’s JNJ-78436735, CanSino Biologics’ Convidicea (AD5-nCoV) and Russia’s Sputnik V, AZD1222 are all examples of this type. So what kind of vaccine is it? How does it work?
Like all vaccines, this method is designed to trick our body into getting infected. These homemade spike proteins will train our bodies to detect and terminate any actual SARS-CoV-2 infection before the virus causes severe damage. The adenovirus vector COVID-19 vaccine constructs the gene of S protein of SARS-CoV-2 into the adenovirus genome, and the shell is still the normal shell protein of the adenovirus. Therefore, when adenovirus infects host cells, the genes encoding the SARS-CoV-2 S protein are released to the host cell, and S protein is synthesized in the cytoplasm, which stimulates a series of immune responses.
The advantage of an adenovirus-based vaccine is that it can induce humoral and cellular immunity at the same time. Humoral immunity produces antibodies that bind to viruses, preventing viruses from entering human cells, that’s to say, viruses are recognized, swallowed, and degraded by macrophages outside the cells. For viruses that have been lucky enough to enter cells, they can be recognized by cellular immune mechanisms and cytokine secreted by killer T cells to lyse the infected cells.
0 notes
Photo

Packing of DNA Double Helices Into Chromatids The length of human DNA molecules far exceeds the diameter of the cell nucleus. DNA is compacted into orderly structures ranging from nucleosomes to metaphase chromatids. In the nucleus, DNA is folded into nucleosomes, which in turn are part of increasingly higher orders of folding. The greatest degree of DNA compaction is needed for cell division. The longest human chromosome (chromosome 1) contains about 246 million base pairs and has a length ~15,000 times the diameter of a typical nucleus. The organization of DNA also affects the transcription of genes. The basic unit of folding is the nucleosome, of which several types exist. Nucleosomes contain a core particle that consists of eight histone proteins, a DNA helix of ~147 base pairs that encircles the histones ~1.7 times, and linker DNA of ~40 base pairs to which histone H1 is often bound. N- and C-terminal tails of the histones protrude from nucleosome core particles. Certain amino acids in these histone tails can be modified. The resulting structure of the histone tails affects the packing, replication, and transcription of DNA https://www.instagram.com/p/CnXatjYrXOH/?igshid=NGJjMDIxMWI=
0 notes
Text
Which wants to become a scientist/who is actually granted in scientific disciplines? Scientific disciplines identity being a contact lens for you to exploring the governmental measurement from the nature associated with technology.
Within this study, ultrasound examination Radio wave signals were received during HIFU direct exposure. Any link coefficient was utilized to judge the changes happening in the Radiation alerts backscattered through the energy sore. In addition, the stop corresponding algorithm has become performed to make up your tissues action through HIFU direct exposure. The new final results reveal that link coefficients within the key spot reduced substantially using HIFU exposure, indicating the backscattered Radio wave signs modified because of muscle coagulation. (H) This year Your Asia Modern society regarding Used ScienceObjectiveA main goal inside the treatment of recent joint disease is the protection against mutual destruction. The need for radiographic further advancement in the first year pertaining to guessing even more radiographic further advancement has not been looked at comparatively with standard predictive components. MethodsPatients along with arthritis associated with smaller compared to Half a dozen months' period have been contained in the prospective France ESPOIR cohort. Radiographs were acquired and also changed Razor-sharp scores had been determined by any blinded readers. The interest rate involving development was determined within the fresh, then over the subsequent along with 3rd decades. Quick progression ended up being looked as a new bigger as compared to 5-point twelve-monthly rise in the whole Sharpened report. ResultsIn complete, Five hundred individuals acquired comprehensive data offered after Three years as well as had been included. The whole Razor-sharp score suggested rapid development inside 123 people (25%) in 12 months One and also 80 patients (18%) in years 2/3. By simply logistic regression, your parameters on their own related to rapid further advancement in years 2/3 ended up calendar year A single rapid AG 013736 clinical trial advancement of your erosion and total Sharpened standing, basic break down Razor-sharp report, the actual serologic U . s . School associated with Rheumatology/European League Versus Rheumatism qualification, and interleukin-6 degree. Any time these kinds of variables have been combined, 12 months One particular speedy further advancement created the greatest info to be able to projecting many years 2/3 rapid advancement. ConclusionFirst-year radiologic advancement is the greatest unbiased forecaster associated with even more rapid progression in early joint disease.Esophageal atresia is a type of as well as life-threatening delivery deficiency using a inadequately comprehended etiology. On this examine, we all analyzed the succession variants associated with coding areas for any group of esophageal atresia-related body's genes which include MYCN, SOX2, CHD7, GLI3, FGFR2 and also PTEN for variations making use of PCR-based targeted enrichment and next-generation sequencing inside 27 patients along with esophageal atresia. Genomic replicate amount alternative investigation has been done using Affymetrix SNP Six.3. We all discovered a delaware novo heterozygous mutation within the N-terminal area of the GLI3 gene (h.332 Capital t bigger as compared to H, p.M111T) within a individual along with esophageal atresia and also hemivertebrae. The actual N-terminal location (aminos 1-397) involving GLI3 provides the repressor site, that communicates using Skiing loved ones healthy proteins.
1 note
·
View note