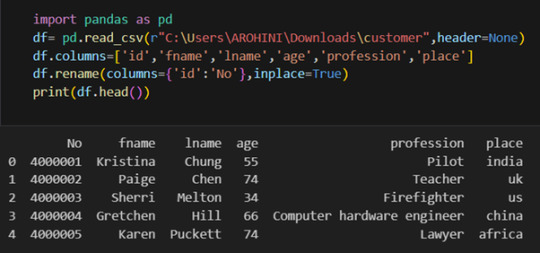
#rename columns in pandas dataframe
Text
DataFrame in Pandas: Guide to Creating Awesome DataFrames
Explore how to create a dataframe in Pandas, including data input methods, customization options, and practical examples.
Data analysis used to be a daunting task, reserved for statisticians and mathematicians. But with the rise of powerful tools like Python and its fantastic library, Pandas, anyone can become a data whiz! Pandas, in particular, shines with its DataFrames, these nifty tables that organize and manipulate data like magic. But where do you start? Fear not, fellow data enthusiast, for this guide will…
View On WordPress
#advanced dataframe features#aggregating data in pandas#create dataframe from dictionary in pandas#create dataframe from list in pandas#create dataframe in pandas#data manipulation in pandas#dataframe indexing#filter dataframe by condition#filter dataframe by multiple conditions#filtering data in pandas#grouping data in pandas#how to make a dataframe in pandas#manipulating data in pandas#merging dataframes#pandas data structures#pandas dataframe tutorial#python dataframe basics#rename columns in pandas dataframe#replace values in pandas dataframe#select columns in pandas dataframe#select rows in pandas dataframe#set column names in pandas dataframe#set row names in pandas dataframe
0 notes
Text
Beginner’s Guide: Data Analysis with Pandas
Data analysis is the process of sorting through all the data, looking for patterns, connections, and interesting things. It helps us make sense of information and use it to make decisions or find solutions to problems. When it comes to data analysis and manipulation in Python, the Pandas library reigns supreme. Pandas provide powerful tools for working with structured data, making it an indispensable asset for both beginners and experienced data scientists.
What is Pandas?
Pandas is an open-source Python library for data manipulation and analysis. It is built on top of NumPy, another popular numerical computing library, and offers additional features specifically tailored for data manipulation and analysis. There are two primary data structures in Pandas:
• Series: A one-dimensional array capable of holding any type of data.
• DataFrame: A two-dimensional labeled data structure similar to a table in relational databases.
It allows us to efficiently process and analyze data, whether it comes from any file types like CSV files, Excel spreadsheets, SQL databases, etc.
How to install Pandas?
We can install Pandas using the pip command. We can run the following codes in the terminal.
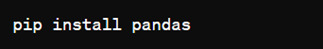
After installing, we can import it using:

How to load an external dataset using Pandas?
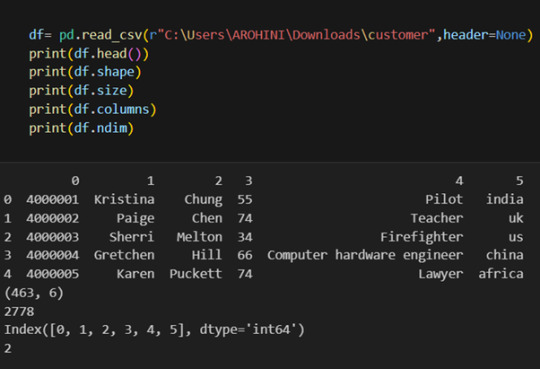
Pandas provide various functions for loading data into a data frame. One of the most commonly used functions is pd.read_csv() for reading CSV files. For example:
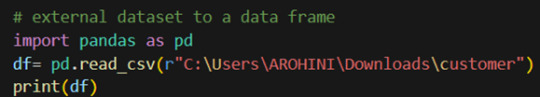
The output of the above code is:
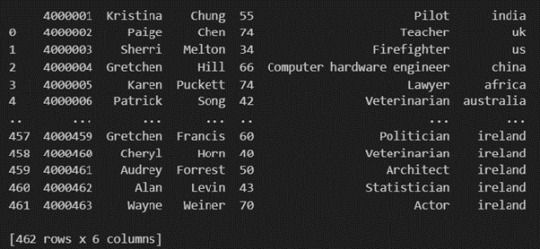
Once your data is loaded into a data frame, you can start exploring it. Pandas offers numerous methods and attributes for getting insights into your data. Here are a few examples:
df.head(): View the first few rows of the DataFrame.
df.tail(): View the last few rows of the DataFrame.
http://df.info(): Get a concise summary of the DataFrame, including data types and missing values.
df.describe(): Generate descriptive statistics for numerical columns.
df.shape: Get the dimensions of the DataFrame (rows, columns).
df.columns: Access the column labels of the DataFrame.
df.dtypes: Get the data types of each column.
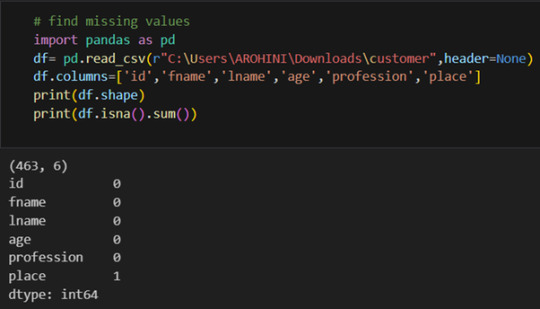
In data analysis, it is essential to do data cleaning. Pandas provide powerful tools for handling missing data, removing duplicates, and transforming data. Some common data-cleaning tasks include:
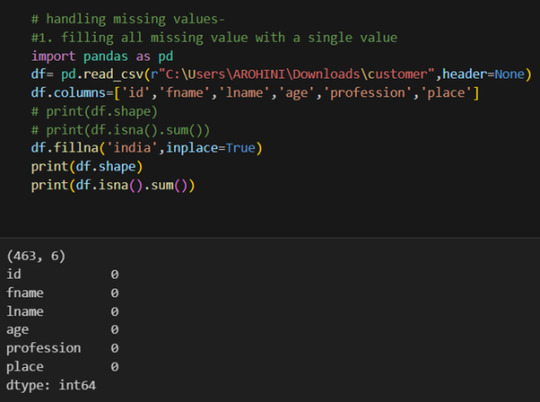
Handling missing values using methods like df.dropna() or df.fillna().
Removing duplicate rows with df.drop_duplicates().
Data type conversion using df.astype().
Renaming columns with df.rename().
Pandas excels in data manipulation tasks such as selecting subsets of data, filtering rows, and creating new columns. Here are a few examples:
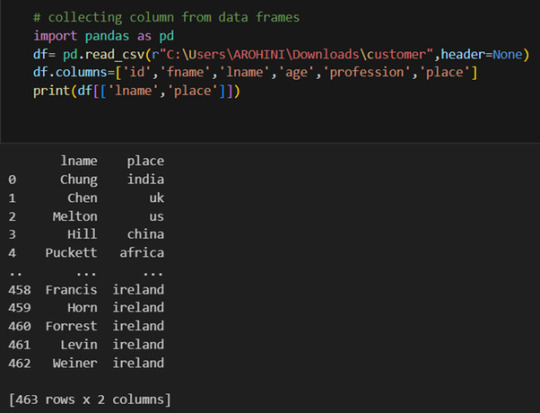
Selecting columns: df[‘column_name’] or df[[‘column1’, ‘column2’]].
Filtering rows based on conditions: df[df[‘column’] > value].
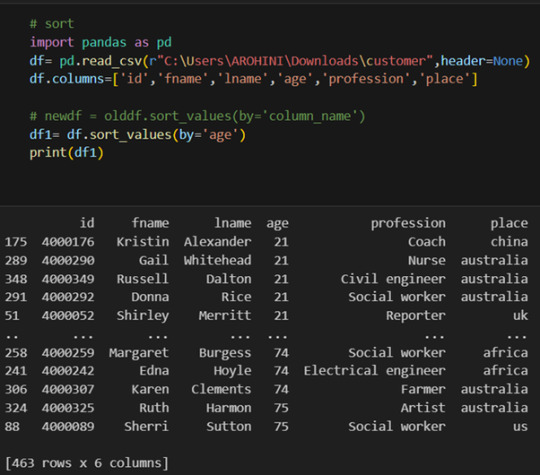
Sorting data: df.sort_values(by=’column’).
Grouping data: df.groupby(‘column’).mean().
With data cleaned and prepared, you can use Pandas to perform various analyses. Whether you’re computing statistics, performing exploratory data analysis, or building predictive models, Pandas provides the tools you need. Additionally, Pandas integrates seamlessly with other libraries such as Matplotlib and Seaborn for data visualization
#data analytics#panda#business analytics course in kochi#cybersecurity#data analytics training#data analytics course in kochi#data analytics course
0 notes
Text
KMeans Clustering Assignment
Import the modules
from pandas import Series, DataFrame
import pandas as pd
import numpy as np
import matplotlib.pylab as plt
from sklearn.model_selection import train_test_split
from sklearn import preprocessing
from sklearn.cluster import KMeans
Load the dataset
data = pd.read_csv("C:\Users\guy3404\OneDrive - MDLZ\Documents\Cross Functional Learning\AI COP\Coursera\machine_learning_data_analysis\Datasets\tree_addhealth.csv")
data.head()
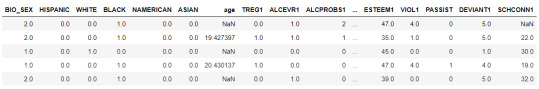
upper-case all DataFrame column names
data.columns = map(str.upper, data.columns)
Data Management
data_clean = data.dropna()
data_clean.head()
subset clustering variables
cluster=data_clean[['ALCEVR1','MAREVER1','ALCPROBS1','DEVIANT1','VIOL1',
'DEP1','ESTEEM1','SCHCONN1','PARACTV', 'PARPRES','FAMCONCT']]
cluster.describe()

standardize clustering variables to have mean=0 and sd=1
clustervar=cluster.copy()
clustervar['ALCEVR1']=preprocessing.scale(clustervar['ALCEVR1'].astype('float64'))
clustervar['ALCPROBS1']=preprocessing.scale(clustervar['ALCPROBS1'].astype('float64'))
clustervar['MAREVER1']=preprocessing.scale(clustervar['MAREVER1'].astype('float64'))
clustervar['DEP1']=preprocessing.scale(clustervar['DEP1'].astype('float64'))
clustervar['ESTEEM1']=preprocessing.scale(clustervar['ESTEEM1'].astype('float64'))
clustervar['VIOL1']=preprocessing.scale(clustervar['VIOL1'].astype('float64'))
clustervar['DEVIANT1']=preprocessing.scale(clustervar['DEVIANT1'].astype('float64'))
clustervar['FAMCONCT']=preprocessing.scale(clustervar['FAMCONCT'].astype('float64'))
clustervar['SCHCONN1']=preprocessing.scale(clustervar['SCHCONN1'].astype('float64'))
clustervar['PARACTV']=preprocessing.scale(clustervar['PARACTV'].astype('float64'))
clustervar['PARPRES']=preprocessing.scale(clustervar['PARPRES'].astype('float64'))
split data into train and test sets
clus_train, clus_test = train_test_split(clustervar, test_size=.3, random_state=123)
k-means cluster analysis for 1-9 clusters
from scipy.spatial.distance import cdist
clusters=range(1,10)
meandist=[]
for k in clusters:
model=KMeans(n_clusters=k)
model.fit(clus_train)
clusassign=model.predict(clus_train)
meandist.append(sum(np.min(cdist(clus_train, model.cluster_centers_, 'euclidean'), axis=1))
/ clus_train.shape[0])
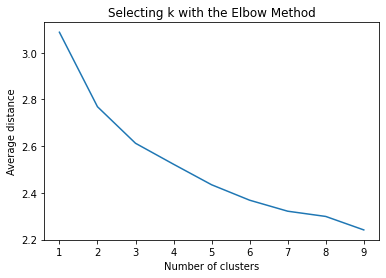
"""
Plot average distance from observations from the cluster centroid
to use the Elbow Method to identify number of clusters to choose
"""
plt.plot(clusters, meandist)
plt.xlabel('Number of clusters')
plt.ylabel('Average distance')
plt.title('Selecting k with the Elbow Method')
Interpret 3 cluster solution
model3=KMeans(n_clusters=3)
model3.fit(clus_train)
clusassign=model3.predict(clus_train)
plot clusters
from sklearn.decomposition import PCA
pca_2 = PCA(2)
plot_columns = pca_2.fit_transform(clus_train)
plt.scatter(x=plot_columns[:,0], y=plot_columns[:,1], c=model3.labels_,)
plt.xlabel('Canonical variable 1')
plt.ylabel('Canonical variable 2')
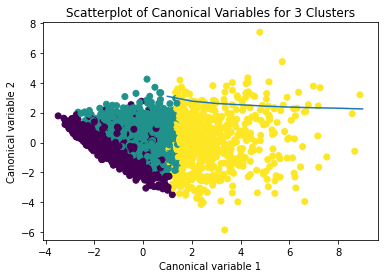
plt.title('Scatterplot of Canonical Variables for 3 Clusters')
plt.show()
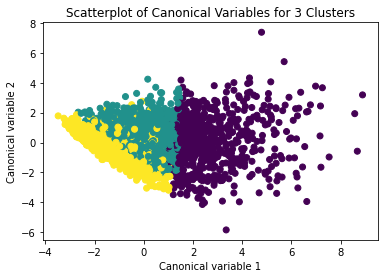
The datapoints of the 2 clusters in the left are less spread out but have more overlaps. The cluster to the right is more distinct but has more spread in the data points
"""
BEGIN multiple steps to merge cluster assignment with clustering variables to examine
cluster variable means by cluster
"""
create a unique identifier variable from the index for the
cluster training data to merge with the cluster assignment variable
clus_train.reset_index(level=0, inplace=True)
create a list that has the new index variable
cluslist=list(clus_train['index'])
create a list of cluster assignments
labels=list(model3.labels_)
combine index variable list with cluster assignment list into a dictionary
newlist=dict(zip(cluslist, labels))
newlist
convert newlist dictionary to a dataframe
newclus=DataFrame.from_dict(newlist, orient='index')
newclus
rename the cluster assignment column
newclus.columns = ['cluster']
now do the same for the cluster assignment variable
create a unique identifier variable from the index for the
cluster assignment dataframe
to merge with cluster training data
newclus.reset_index(level=0, inplace=True)
merge the cluster assignment dataframe with the cluster training variable dataframe
by the index variable
merged_train=pd.merge(clus_train, newclus, on='index')
merged_train.head(n=100)
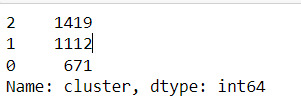
cluster frequencies
merged_train.cluster.value_counts()
"""
END multiple steps to merge cluster assignment with clustering variables to examine
cluster variable means by cluster
"""
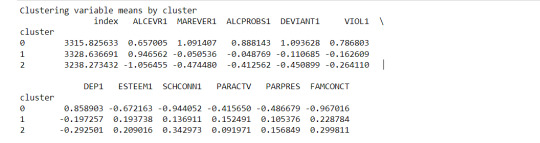
FINALLY calculate clustering variable means by cluster
clustergrp = merged_train.groupby('cluster').mean()
print ("Clustering variable means by cluster")
print(clustergrp)
validate clusters in training data by examining cluster differences in GPA using ANOVA
first have to merge GPA with clustering variables and cluster assignment data
gpa_data=data_clean['GPA1']
split GPA data into train and test sets
gpa_train, gpa_test = train_test_split(gpa_data, test_size=.3, random_state=123)
gpa_train1=pd.DataFrame(gpa_train)
gpa_train1.reset_index(level=0, inplace=True)
merged_train_all=pd.merge(gpa_train1, merged_train, on='index')
sub1 = merged_train_all[['GPA1', 'cluster']].dropna()
Print statistical summary by cluster
import statsmodels.formula.api as smf
import statsmodels.stats.multicomp as multi
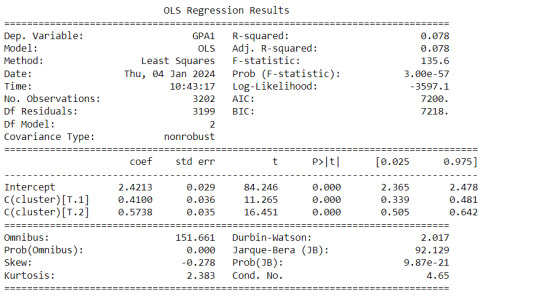
gpamod = smf.ols(formula='GPA1 ~ C(cluster)', data=sub1).fit()
print (gpamod.summary())
print ('means for GPA by cluster')
m1= sub1.groupby('cluster').mean()
print (m1)
print ('standard deviations for GPA by cluster')
m2= sub1.groupby('cluster').std()
print (m2)

Interpretation
The clustering average summary shows Cluster 0 has higher alcohol and marijuana problems, shows higher deviant and violent behavior, suffers from depression, has low self esteem,school connectedness, paraental and family connectedness. On the contrary, Cluster 2 shows the lowest alcohol and marijuana problems, lowest deviant & violent behavior,depression, and higher self esteem,school connectedness, paraental and family connectedness. Further, when validated against GPA score, we observe Cluster 0 shows the lowest average GPA and CLuster 2 has the highest average GPA which aligns with the summary statistics interpretation.
0 notes
Text
rom pandas import Series, DataFrame
import pandas as pd
import numpy as np
import matplotlib.pylab as plt
from sklearn.model_selection import train_test_split
from sklearn import preprocessing
from sklearn.cluster import KMeans
"""
Data Management
"""
data = pd.read_csv("tree_addhealth")
upper-case all DataFrame column names
data.columns = map(str.upper, data.columns)
Data Management
data_clean = data.dropna()
subset clustering variables
cluster=data_clean[['ALCEVR1','MAREVER1','ALCPROBS1','DEVIANT1','VIOL1',
'DEP1','ESTEEM1','SCHCONN1','PARACTV', 'PARPRES','FAMCONCT']]
cluster.describe()
standardize clustering variables to have mean=0 and sd=1
clustervar=cluster.copy()
clustervar['ALCEVR1']=preprocessing.scale(clustervar['ALCEVR1'].astype('float64'))
clustervar['ALCPROBS1']=preprocessing.scale(clustervar['ALCPROBS1'].astype('float64'))
clustervar['MAREVER1']=preprocessing.scale(clustervar['MAREVER1'].astype('float64'))
clustervar['DEP1']=preprocessing.scale(clustervar['DEP1'].astype('float64'))
clustervar['ESTEEM1']=preprocessing.scale(clustervar['ESTEEM1'].astype('float64'))
clustervar['VIOL1']=preprocessing.scale(clustervar['VIOL1'].astype('float64'))
clustervar['DEVIANT1']=preprocessing.scale(clustervar['DEVIANT1'].astype('float64'))
clustervar['FAMCONCT']=preprocessing.scale(clustervar['FAMCONCT'].astype('float64'))
clustervar['SCHCONN1']=preprocessing.scale(clustervar['SCHCONN1'].astype('float64'))
clustervar['PARACTV']=preprocessing.scale(clustervar['PARACTV'].astype('float64'))
clustervar['PARPRES']=preprocessing.scale(clustervar['PARPRES'].astype('float64'))
split data into train and test sets
clus_train, clus_test = train_test_split(clustervar, test_size=.3, random_state=123)
k-means cluster analysis for 1-9 clusters
from scipy.spatial.distance import cdist
clusters=range(1,10)
meandist=[]
for k in clusters:
model=KMeans(n_clusters=k)
model.fit(clus_train)
clusassign=model.predict(clus_train)
meandist.append(sum(np.min(cdist(clus_train, model.cluster_centers_, 'euclidean'), axis=1))
/ clus_train.shape[0])
"""
Plot average distance from observations from the cluster centroid
to use the Elbow Method to identify number of clusters to choose
"""
plt.plot(clusters, meandist)
plt.xlabel('Number of clusters')
plt.ylabel('Average distance')
plt.title('Selecting k with the Elbow Method')
Interpret 3 cluster solution
model3=KMeans(n_clusters=3)
model3.fit(clus_train)
clusassign=model3.predict(clus_train)
plot clusters
from sklearn.decomposition import PCA
pca_2 = PCA(2)
plot_columns = pca_2.fit_transform(clus_train)
plt.scatter(x=plot_columns[:,0], y=plot_columns[:,1], c=model3.labels_,)
plt.xlabel('Canonical variable 1')
plt.ylabel('Canonical variable 2')
plt.title('Scatterplot of Canonical Variables for 3 Clusters')
plt.show()
"""
BEGIN multiple steps to merge cluster assignment with clustering variables to examine
cluster variable means by cluster
"""
create a unique identifier variable from the index for the
cluster training data to merge with the cluster assignment variable
clus_train.reset_index(level=0, inplace=True)
create a list that has the new index variable
cluslist=list(clus_train['index'])
create a list of cluster assignments
labels=list(model3.labels_)
combine index variable list with cluster assignment list into a dictionary
newlist=dict(zip(cluslist, labels))
newlist
convert newlist dictionary to a dataframe
newclus=DataFrame.from_dict(newlist, orient='index')
newclus
rename the cluster assignment column
newclus.columns = ['cluster']
now do the same for the cluster assignment variable
create a unique identifier variable from the index for the
cluster assignment dataframe
to merge with cluster training data
newclus.reset_index(level=0, inplace=True)
merge the cluster assignment dataframe with the cluster training variable dataframe
by the index variable
merged_train=pd.merge(clus_train, newclus, on='index')
merged_train.head(n=100)
cluster frequencies
merged_train.cluster.value_counts()
"""
END multiple steps to merge cluster assignment with clustering variables to examine
cluster variable means by cluster
"""
FINALLY calculate clustering variable means by cluster
clustergrp = merged_train.groupby('cluster').mean()
print ("Clustering variable means by cluster")
print(clustergrp)
validate clusters in training data by examining cluster differences in GPA using ANOVA
first have to merge GPA with clustering variables and cluster assignment data
gpa_data=data_clean['GPA1']
split GPA data into train and test sets
gpa_train, gpa_test = train_test_split(gpa_data, test_size=.3, random_state=123)
gpa_train1=pd.DataFrame(gpa_train)
gpa_train1.reset_index(level=0, inplace=True)
merged_train_all=pd.merge(gpa_train1, merged_train, on='index')
sub1 = merged_train_all[['GPA1', 'cluster']].dropna()
import statsmodels.formula.api as smf
import statsmodels.stats.multicomp as multi
gpamod = smf.ols(formula='GPA1 ~ C(cluster)', data=sub1).fit()
print (gpamod.summary())
print ('means for GPA by cluster')
m1= sub1.groupby('cluster').mean()
print (m1)
print ('standard deviations for GPA by cluster')
m2= sub1.groupby('cluster').std()
print (m2)
mc1 = multi.MultiComparison(sub1['GPA1'], sub1['cluster'])
res1 = mc1.tukeyhsd()
print(res1.summary())
0 notes
Text
Rename Pandas DataFrame Index
I've a csv file without header, with a DateTime index. I want to rename the index and column name, but with df.rename() only the column name is renamed. Bug? I'm on version 0.12.0
In [2]: df = pd.read_csv(r'D:\Data\DataTimeSeries_csv//seriesSM.csv', header=None, parse_dates=[[0]], index_col=[0] )In [3]: df.head()Out[3]: 10 2002-06-18 0.1120002002-06-22 0.1903332002-06-26 0.1340002002-06-30 0.0930002002-07-04 0.098667In [4]: df.rename(index={0:'Date'}, columns={1:'SM'}, inplace=True)In [5]: df.head()Out[5]: SM0 2002-06-18 0.1120002002-06-22 0.1903332002-06-26 0.1340002002-06-30 0.0930002002-07-04 0.098667
https://codehunter.cc/a/python/rename-pandas-dataframe-index
0 notes
Text
Machine Learning for Data Analysis - Week 4
#Load the data and convert the variables to numeric
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LassoLarsCV
import statsmodels.formula.api as smf
import statsmodels.stats.multicomp as multi
from sklearn import preprocessing
from sklearn.cluster import KMeans
data = pd.read_csv('gapminder.csv', low_memory=False)
data['urbanrate'] = pd.to_numeric(data['urbanrate'], errors='coerce')
data['incomeperperson'] = pd.to_numeric(data['incomeperperson'], errors='coerce')
data['femaleemployrate'] = pd.to_numeric(data['femaleemployrate'], errors='coerce')
data['breastcancerper100th'] = pd.to_numeric(data['breastcancerper100th'], errors='coerce')
data['internetuserate'] = pd.to_numeric(data['internetuserate'], errors='coerce')
data['employrate'] = pd.to_numeric(data['employrate'], errors='coerce')
data['polityscore'] = pd.to_numeric(data['polityscore'], errors='coerce')
data['lifeexpectancy'] = pd.to_numeric(data['lifeexpectancy'], errors='coerce')
sub1 = data.copy()
data_clean = sub1.dropna()
#Subset the clustering variables
cluster = data_clean[['incomeperperson','femaleemployrate','breastcancerper100th','internetuserate',
'employrate', 'polityscore', 'lifeexpectancy']]
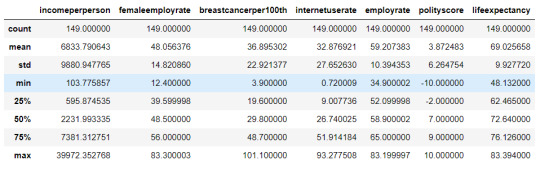
cluster.describe()
#Standardize the clustering variables to have mean = 0 and standard deviation = 1
clustervar=cluster.copy()
clustervar['incomeperperson']=preprocessing.scale(clustervar['incomeperperson'].astype('float64'))
clustervar['femaleemployrate']=preprocessing.scale(clustervar['femaleemployrate'].astype('float64'))
clustervar['breastcancerper100th']=preprocessing.scale(clustervar['breastcancerper100th'].astype('float64'))
clustervar['internetuserate']=preprocessing.scale(clustervar['internetuserate'].astype('float64'))
clustervar['employrate']=preprocessing.scale(clustervar['employrate'].astype('float64'))
clustervar['polityscore']=preprocessing.scale(clustervar['polityscore'].astype('float64'))
clustervar['lifeexpectancy']=preprocessing.scale(clustervar['lifeexpectancy'].astype('float64'))
#Split the data into train and test sets
clus_train, clus_test = train_test_split(clustervar, test_size=.3, random_state=123)
#Perform k-means cluster analysis for 1-9 clusters
from scipy.spatial.distance import cdist
clusters = range(1,10)
meandist = []
for k in clusters:
model = KMeans(n_clusassign = k)
model.fit(clus_train)
clusters = model.predict(clus_train)
meandist.append(sum(np.min(cdist(clus_train, model.cluster_centers_, 'euclidean'), axis=1))
/ clus_train.shape[0])
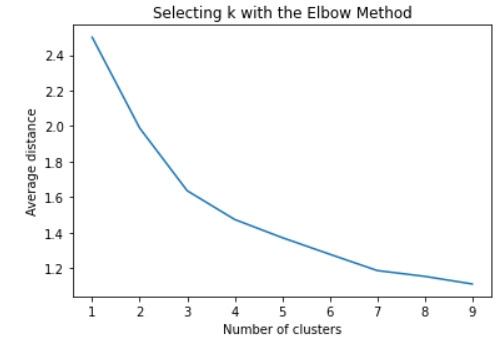
#Plot average distance from observations from the cluster centroid to use the Elbow Method to identify number of clusters to choose
plt.plot(clusters, meandist)
plt.xlabel('Number of clusters')
plt.ylabel('Average distance')
plt.title('Selecting k with the Elbow Method')
plt.show()
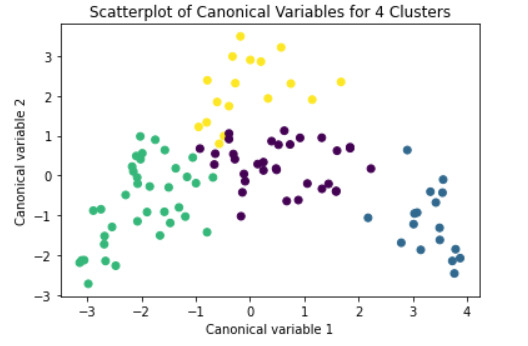
#Interpret 3 cluster solution
model3 = KMeans(n_clusters=4)
model3.fit(clus_train)
clusassign = model3.predict(clus_train)
#Plot the clusters
from sklearn.decomposition import PCA
pca_2 = PCA(2)
plt.figure()
plot_columns = pca_2.fit_transform(clus_train)
plt.scatter(x=plot_columns[:,0], y=plot_columns[:,1], c=model3.labels_,)
plt.xlabel('Canonical variable 1')
plt.ylabel('Canonical variable 2')
plt.title('Scatterplot of Canonical Variables for 4 Clusters')
plt.show()
#Create a unique identifier variable from the index for the cluster training data to merge with the cluster assignment variable.
clus_train.reset_index(level=0, inplace=True)
#Create a list that has the new index variable
cluslist = list(clus_train['index'])
#Create a list of cluster assignments
labels = list(model3.labels_)
#Combine index variable list with cluster assignment list into a dictionary
newlist = dict(zip(cluslist, labels))
print(newlist)

#Convert newlist dictionary to a dataframe
newclus = pd.DataFrame.from_dict(newlist, orient='index')
#Rename the cluster assignment column
newclus.columns = ['cluster']
newclus

#Create a unique identifier variable from the index for the cluster assignment dataframe to merge with cluster training data
newclus.reset_index(level=0, inplace=True)
#Merge the cluster assignment dataframe with the cluster training variable dataframe by the index variable
merged_train = pd.merge(clus_train, newclus, on='index')
merged_train.head(n=100)
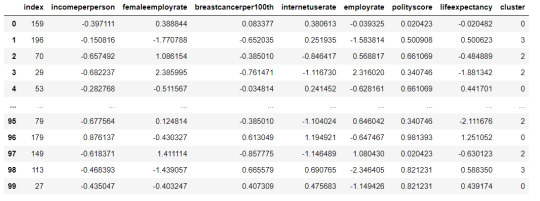
#Cluster frequencies
merged_train.cluster.value_counts()
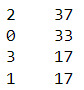
#Calculate clustering variable means by cluster
clustergrp = merged_train.groupby('cluster').mean()
print ("Clustering variable means by cluster")
clustergrp
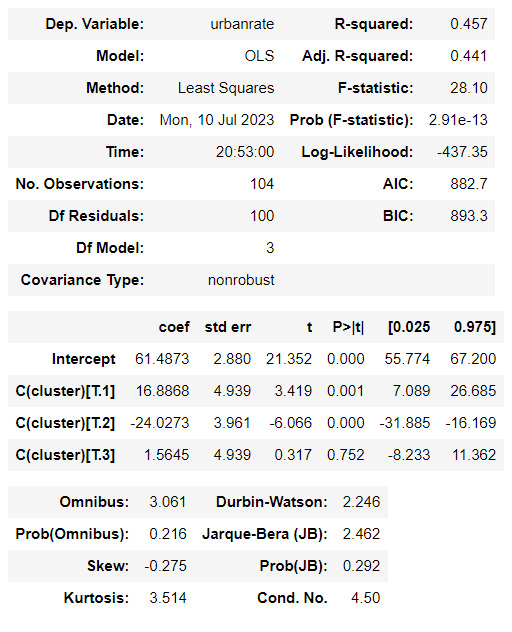
#Validate clusters in training data by examining cluster differences in urbanrate using ANOVA.
#First, merge urbanrate with clustering variables and cluster assignment data
urbanrate_data = data_clean['urbanrate']
#Split urbanrate data into train and test sets
urbanrate_train, internetuserate_test = train_test_split(urbanrate_data, test_size=.3, random_state=123)
urbanrate_train1=pd.DataFrame(urbanrate_train)
urbanrate_train1.reset_index(level=0, inplace=True)
merged_train_all=pd.merge(urbanrate_train1, merged_train, on='index')
sub5 = merged_train_all[['urbanrate', 'cluster']].dropna()
urbanrate_mod = smf.ols(formula='urbanrate ~ C(cluster)', data=sub5).fit()
urbanrate_mod.summary()
#Means for urbanrate by cluster
m1= sub5.groupby('cluster').mean()
m1
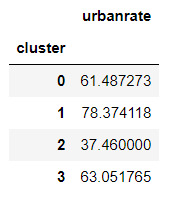
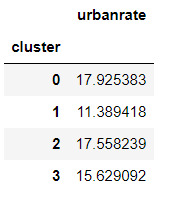
#Standard deviations for urbanrate by cluster
m2= sub5.groupby('cluster').std()
m2
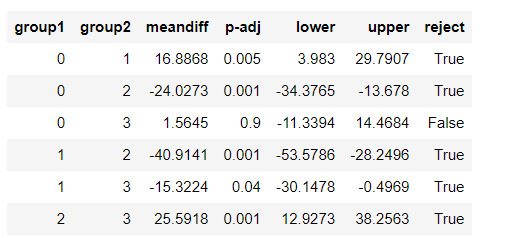
mc1 = multi.MultiComparison(sub5['urbanrate'], sub5['cluster'])
res1 = mc1.tukeyhsd()
res1.summary()
0 notes
Text
Código K- means craters of mars
-- coding: utf-8 --
"""
Created on Fri Jun 16 19:08:39 2023
@author: ANGELA
"""
from pandas import Series, DataFrame
import pandas
import numpy as np
import matplotlib.pylab as plt
from sklearn.model_selection import train_test_split
from sklearn import preprocessing
from sklearn.cluster import KMeans
"""Data management"""
data = pandas.read_csv('marscrater_pds.csv', low_memory=False)
data['LATITUDE_CIRCLE_IMAGE']=pandas.to_numeric(data['LATITUDE_CIRCLE_IMAGE'],errors='coerce')
data['LONGITUDE_CIRCLE_IMAGE']=pandas.to_numeric(data['LONGITUDE_CIRCLE_IMAGE'],errors='coerce')
data['DIAM_CIRCLE_IMAGE']=pandas.to_numeric(data['DIAM_CIRCLE_IMAGE'],errors='coerce')
data['NUMBER_LAYERS']=pandas.to_numeric(data['NUMBER_LAYERS'],errors='coerce')
data['DEPTH_RIMFLOOR_TOPOG']=pandas.to_numeric(data['DEPTH_RIMFLOOR_TOPOG'],errors='coerce')
upper-case all DataFrame column names
data.columns = map(str.upper, data.columns)
data_clean =data.dropna()
target = data_clean.DEPTH_RIMFLOOR_TOPOG
select predictor variables and target variable as separate data sets
cluster= data_clean[['LATITUDE_CIRCLE_IMAGE','LONGITUDE_CIRCLE_IMAGE','DIAM_CIRCLE_IMAGE']]
cluster.describe()
standardize clustering variables to have mean=0 and sd=1
clustervar=cluster.copy()
clustervar['LATITUDE_CIRCLE_IMAGE']=preprocessing.scale(clustervar['LATITUDE_CIRCLE_IMAGE'].astype('float64'))
clustervar['LONGITUDE_CIRCLE_IMAGE']=preprocessing.scale(clustervar['LONGITUDE_CIRCLE_IMAGE'].astype('float64'))
clustervar['DIAM_CIRCLE_IMAGE']=preprocessing.scale(clustervar['DIAM_CIRCLE_IMAGE'].astype('float64'))
clustervar['DEPTH_RIMFLOOR_TOPOG']=preprocessing.scale(clustervar['DEPTH_RIMFLOOR_TOPOG'].astype('float64'))
split data into train and test sets
clus_train, clus_test = train_test_split(clustervar, test_size=.3, random_state=200)
k-means cluster analysis for 1-9 clusters
from scipy.spatial.distance import cdist
clusters=range(1,10)
meandist=[]
Calculate cluster
for k in clusters:
model=KMeans(n_clusters=k)
model.fit(clus_train)
clusassign=model.predict(clus_train)
meandist.append(sum(np.min(cdist(clus_train, model.cluster_centers_, 'euclidean'), axis=1))
/ clus_train.shape[0])
"""
Plot average distance from observations from the cluster centroid
to use the Elbow Method to identify number of clusters to choose
"""
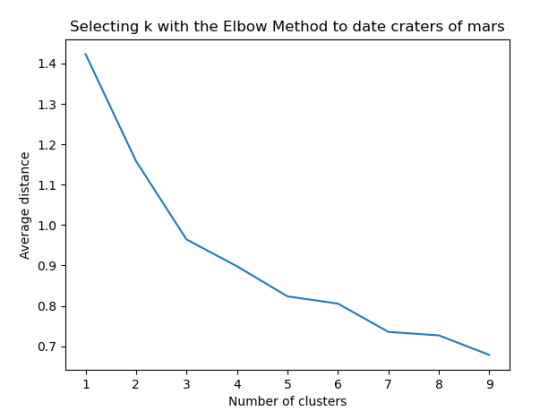
plt.plot(clusters, meandist)
plt.xlabel('Number of clusters')
plt.ylabel('Average distance')
plt.title('Selecting k with the Elbow Method to date craters of mars')
Interpret 3 cluster solution
model3=KMeans(n_clusters=3)
model3.fit(clus_train)
clusassign=model3.predict(clus_train)
plot clusters
from sklearn.decomposition import PCA
pca_2 = PCA(2)
plot_columns = pca_2.fit_transform(clus_train)
plt.scatter(x=plot_columns[:,0], y=plot_columns[:,1], c=model3.labels_,)
plt.xlabel('Canonical variable 1')
plt.ylabel('Canonical variable 2')
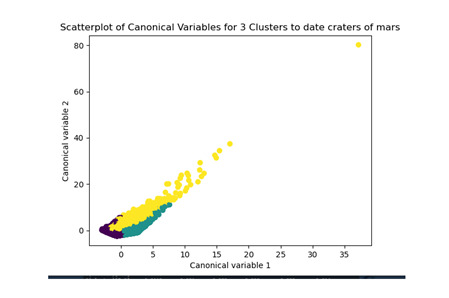
plt.title('Scatterplot of Canonical Variables for 3 Clusters to date craters of mars')
plt.show()
"""
BEGIN multiple steps to merge cluster assignment with clustering variables to examine
cluster variable means by cluster
"""
create a unique identifier variable from the index for the
cluster training data to merge with the cluster assignment variable
clus_train.reset_index(level=0, inplace=True)
create a list that has the new index variable
cluslist=list(clus_train['index'])
create a list of cluster assignments
labels=list(model3.labels_)
combine index variable list with cluster assignment list into a dictionary
newlist=dict(zip(cluslist, labels))
newlist
convert newlist dictionary to a dataframe
newclus=DataFrame.from_dict(newlist, orient='index')
newclus
rename the cluster assignment column
newclus.columns = ['cluster']
now do the same for the cluster assignment variable
create a unique identifier variable from the index for the
cluster assignment dataframe
to merge with cluster training data
newclus.reset_index(level=0, inplace=True)
merge the cluster assignment dataframe with the cluster training variable dataframe
by the index variable
merged_train=pandas.merge(clus_train, newclus, on='index')
merged_train.head(n=100)
cluster frequencies
merged_train.cluster.value_counts()
"""
END multiple steps to merge cluster assignment with clustering variables to examine
cluster variable means by cluster
"""
FINALLY calculate clustering variable means by cluster
clustergrp = merged_train.groupby('cluster').mean()
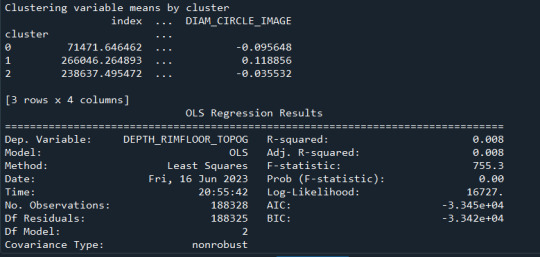
print ("Clustering variable means by cluster")
print(clustergrp)
validate clusters in training data by examining cluster differences in DEPTH_RIMFLOOR_TOPOG using ANOVA
first have to merge DEPTH_RIMFLOOR_TOPOG with clustering variables and cluster assignment data
DRT_data=data_clean['DEPTH_RIMFLOOR_TOPOG']
split DEPTH_RIMFLOOR_TOPOG data into train and test sets
DRT_train, DRT_test = train_test_split(DRT_data, test_size=.3, random_state=123)
DRT_train1=pandas.DataFrame(DRT_train)
DRT_train1.reset_index(level=0, inplace=True)
merged_train_all=pandas.merge(DRT_train1, merged_train, on='index')
sub1 = merged_train_all[['DEPTH_RIMFLOOR_TOPOG', 'cluster']].dropna()
import statsmodels.formula.api as smf
import statsmodels.stats.multicomp as multi
DTRmod = smf.ols(formula='DEPTH_RIMFLOOR_TOPOG ~ C(cluster)', data=sub1).fit()
print (DTRmod.summary())
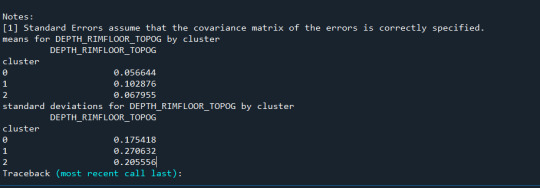
print ('means for DEPTH_RIMFLOOR_TOPOG by cluster')
m1= sub1.groupby('cluster').mean()
print (m1)
print ('standard deviations for DEPTH_RIMFLOOR_TOPOG by cluster')
m2= sub1.groupby('cluster').std()
print (m2)
mc1 = multi.MultiComparison(sub1['GPA1'], sub1['cluster'])
res1 = mc1.tukeyhsd()
print(res1.summary())
Resultados
Los cluster posibles son 3, 5, 7 y 8 donde se evidencia un punto de quiebre en la ilustración, por lo que se realizó la prueba con 3 clusters y se evidencia que puede presentarse un overfiting al esta muy cerca y cobre puestos los clusters

0 notes
Text
Data Management Decisions
STEP 1: Data Management Decisions
Coding out missing data: Identify how missing data is coded in your dataset and decide on a specific value to represent missing data. Let's say missing data is coded as "999" in your dataset, and you decide to recode it as "NA" to indicate missing values.
Coding in valid data: Ensure that valid data is appropriately coded and labeled in your dataset. Check if there are any inconsistencies or errors in the data coding and correct them if necessary.
Recoding variables: If needed, recode variables to align with your research question. For example, if you have a variable "Age" and want to create age groups, you can recode it into categories like "18-24," "25-34," "35-44," and so on.
Creating secondary variables: If there are specific calculations or derived variables that would be useful for your analysis, create them based on existing variables. For instance, if you have variables for height and weight, you can calculate the body mass index (BMI) as a secondary variable.
STEP 2: Running Frequency Distributions
Once you have implemented your data management decisions, you can proceed to run frequency distributions for your chosen variables. Ensure that your output is organized, labeled, and easy to interpret.
Here's an example program in Python to demonstrate the process:
import pandas as pd
Assuming you have a DataFrame called 'data' containing your variables
Code out missing data
data.replace(999, pd.NA, inplace=True)
Recode variables
data['Age_Group'] = pd.cut(data['Age'], bins=[18, 25, 35, 45, 55], labels=['18-24', '25-34', '35-44', '45-54'])
Create secondary variable
data['BMI'] = data['Weight'] / ((data['Height'] / 100) ** 2)
Frequency distribution for variable 'Gender'
gender_freq = data['Gender'].value_counts().reset_index().rename(columns={'index': 'Gender', 'Gender': 'Frequency'})
Frequency distribution for variable 'Age_Group'
age_group_freq = data['Age_Group'].value_counts().reset_index().rename(columns={'index': 'Age_Group', 'Age_Group': 'Frequency'})
Frequency distribution for variable 'BMI'
bmi_freq = data['BMI'].value_counts().reset_index().rename(columns={'index': 'BMI', 'BMI': 'Frequency'})
Print the frequency distributions
print("Frequency Distribution - Gender:")
print(gender_freq)
print()
print("Frequency Distribution - Age Group:")
print(age_group_freq)
print()
print("Frequency Distribution - BMI:")
print(bmi_freq)
0 notes
Text
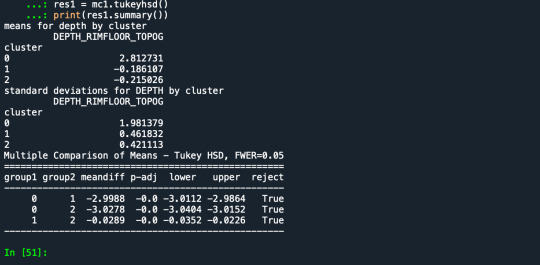
MOD 4
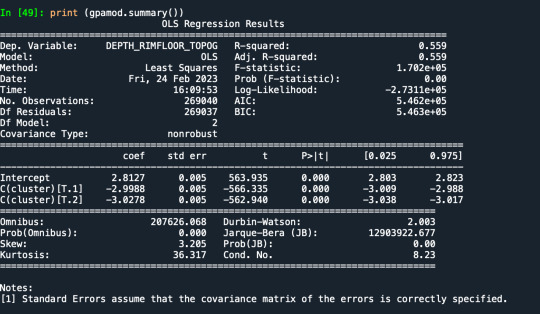
Results from the OLS regression results using the depth rim floor as the variable. the numbers appear to be close and now too far off. Cluster 0 having the biggest depth rim floor to the top.
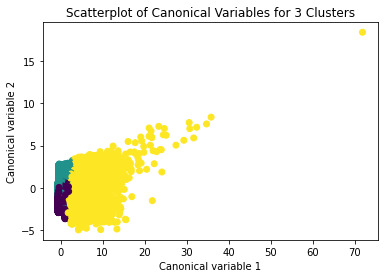
demonstrates that there is a giant overlap within the cluster groups however the yellow group appears to be more dispersed.
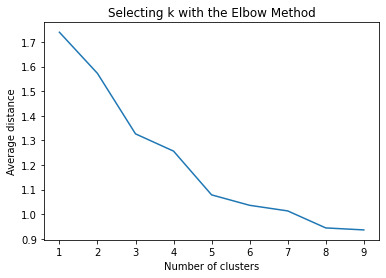
shows that theyre may be overlaps values 2, 3, 4, 5,6,7,8. This means that the variables being utilized are similar. Therefore the canonical variable test was performed to reduce the number of available variables.
CODE SCRIPT
from pandas import Series, DataFrame
import pandas as pd
import numpy as np
import matplotlib.pylab as plt
from sklearn.model_selection import train_test_split
from sklearn import preprocessing
from sklearn.cluster import KMeans
"""
Data Management
"""
data = pd.read_csv('https://d3c33hcgiwev3.cloudfront.net/2c7ec69d0edd3b9599c0df80f0901a52_marscrater_pds.csv?Expires=1677369600&Signature=YDtfrRGhpLU3YbElRnuT3BynxPQdU1s3n6D-tR~Kb1tv7gDGdw2cKF49yGsmou3zWhP4ScXqbCGPbSdTd8SCPdZQpGXuj5B9I2lpUXObnn3OWFsNlQDz7WmrsngPFSdWHciEYCpCdYegyMmghimmDw1xZepgByZPuB5-Z6b3fOQ&Key-Pair-Id=APKAJLTNE6QMUY6HBC5A')
upper-case all DataFrame column names
data.columns = map(str.upper, data.columns)
Data Management
data_clean = data.dropna()
data.dtypes
subset clustering variables
cluster=data_clean[['LATITUDE_CIRCLE_IMAGE','LONGITUDE_CIRCLE_IMAGE','DIAM_CIRCLE_IMAGE','DEPTH_RIMFLOOR_TOPOG',
'NUMBER_LAYERS']]
cluster.describe()
standardize clustering variables to have mean=0 and sd=1
clustervar=cluster.copy()
clustervar['LATITUDE_CIRCLE_IMAGE']=preprocessing.scale(clustervar['LATITUDE_CIRCLE_IMAGE'].astype('float64'))
clustervar['LONGITUDE_CIRCLE_IMAGE']=preprocessing.scale(clustervar['LONGITUDE_CIRCLE_IMAGE'].astype('float64'))
clustervar['DIAM_CIRCLE_IMAGE']=preprocessing.scale(clustervar['DIAM_CIRCLE_IMAGE'].astype('float64'))
clustervar['DEPTH_RIMFLOOR_TOPOG']=preprocessing.scale(clustervar['DEPTH_RIMFLOOR_TOPOG'].astype('float64'))
clustervar['NUMBER_LAYERS']=preprocessing.scale(clustervar['NUMBER_LAYERS'].astype('float64'))
split data into train and test sets
clus_train, clus_test = train_test_split(clustervar, test_size=.3, random_state=123)
k-means cluster analysis for 1-9 clusters
from scipy.spatial.distance import cdist
clusters=range(1,10)
meandist=[]
for k in clusters:
model=KMeans(n_clusters=k)
model.fit(clus_train)
clusassign=model.predict(clus_train)
meandist.append(sum(np.min(cdist(clus_train, model.cluster_centers_, 'euclidean'), axis=1))
/ clus_train.shape[0])
"""
Plot average distance from observations from the cluster centroid
to use the Elbow Method to identify number of clusters to choose
"""
plt.plot(clusters, meandist)
plt.xlabel('Number of clusters')
plt.ylabel('Average distance')
plt.title('Selecting k with the Elbow Method')
Interpret 3 cluster solution
model3=KMeans(n_clusters=3)
model3.fit(clus_train)
clusassign=model3.predict(clus_train)
plot clusters
from sklearn.decomposition import PCA
pca_2 = PCA(2)
plot_columns = pca_2.fit_transform(clus_train)
plt.scatter(x=plot_columns[:,0], y=plot_columns[:,1], c=model3.labels_,)
plt.xlabel('Canonical variable 1')
plt.ylabel('Canonical variable 2')
plt.title('Scatterplot of Canonical Variables for 3 Clusters')
plt.show()
"""
BEGIN multiple steps to merge cluster assignment with clustering variables to examine
cluster variable means by cluster
"""
create a unique identifier variable from the index for the
cluster training data to merge with the cluster assignment variable
clus_train.reset_index(level=0, inplace=True)
create a list that has the new index variable
cluslist=list(clus_train['index'])
create a list of cluster assignments
labels=list(model3.labels_)
combine index variable list with cluster assignment list into a dictionary
newlist=dict(zip(cluslist, labels))
newlist
convert newlist dictionary to a dataframe
newclus=DataFrame.from_dict(newlist, orient='index')
newclus
rename the cluster assignment column
newclus.columns = ['cluster']
now do the same for the cluster assignment variable
create a unique identifier variable from the index for the
cluster assignment dataframe
to merge with cluster training data
newclus.reset_index(level=0, inplace=True)
merge the cluster assignment dataframe with the cluster training variable dataframe
by the index variable
merged_train=pd.merge(clus_train, newclus, on='index')
merged_train.head(n=100)
cluster frequencies
merged_train.cluster.value_counts()
"""
END multiple steps to merge cluster assignment with clustering variables to examine
cluster variable means by cluster
"""
FINALLY calculate clustering variable means by cluster
clustergrp = merged_train.groupby('cluster').mean()
print ("Clustering variable means by cluster")
print(clustergrp)
validate clusters in training data by examining cluster differences in GPA using ANOVA
first have to merge GPA with clustering variables and cluster assignment data
depth_data=data_clean['DEPTH_RIMFLOOR_TOPOG']
split GPA data into train and test sets
depth_train, depth_test = train_test_split(depth_data, test_size=.3, random_state=123)
depth_train1=pd.DataFrame(layer_train)
depth_train1.reset_index(level=0, inplace=True)
merged_train_all=pd.merge(depth_train1, merged_train, on='index')
sub1 = merged_train_all[['DEPTH_RIMFLOOR_TOPOG', 'cluster']].dropna()
import statsmodels.formula.api as smf
import statsmodels.stats.multicomp as multi
gpamod = smf.ols(formula='DEPTH_RIMFLOOR_TOPOG ~ C(cluster)', data=sub1).fit()
print (gpamod.summary())
print ('means for depth by cluster')
m1= sub1.groupby('cluster').mean()
print (m1)
print ('standard deviations for DEPTH by cluster')
m2= sub1.groupby('cluster').std()
print (m2)
mc1 = multi.MultiComparison(sub1['DEPTH_RIMFLOOR_TOPOG'], sub1['cluster'])
res1 = mc1.tukeyhsd()
print(res1.summary())
0 notes
Text
import pandas
import numpy
import scipy.stats
import seaborn
import matplotlib.pyplot as plt
nesarc = pandas.read_csv ('nesarc_pds.csv' , low_memory=False)
Set PANDAS to show all columns in DataFrame
pandas.set_option('display.max_columns', None)
#Set PANDAS to show all rows in DataFrame
pandas.set_option('display.max_rows', None)
nesarc.columns = map(str.upper , nesarc.columns)
pandas.set_option('display.float_format' , lambda x:'%f'%x)
#Change my variables to numeric
nesarc['AGE'] = pandas.to_numeric(nesarc['AGE'], errors='coerce') nesarc['S3BQ4'] = pandas.to_numeric(nesarc['S3BQ4'], errors='coerce') nesarc['S3BQ1A5'] = pandas.to_numeric(nesarc['S3BQ1A5'], errors='coerce') nesarc['S3BD5Q2B'] = pandas.to_numeric(nesarc['S3BD5Q2B'], errors='coerce') nesarc['S3BD5Q2E'] = pandas.to_numeric(nesarc['S3BD5Q2E'], errors='coerce') nesarc['MAJORDEP12'] = pandas.to_numeric(nesarc['MAJORDEP12'], errors='coerce') nesarc['GENAXDX12'] = pandas.to_numeric(nesarc['GENAXDX12'], errors='coerce')
#Subset my sample
subset1 = nesarc[(nesarc['AGE']>=18) & (nesarc['AGE']<=30)] # Ages 18-30 subsetc1 = subset1.copy()
subset2 = nesarc[(nesarc['AGE']>=18) & (nesarc['AGE']<=30) & (nesarc['S3BQ1A5']==1)] # Cannabis users, ages 18-30 subsetc2 = subset2.copy()
#Setting missing data for frequency and cannabis use, variables S3BD5Q2E, S3BQ1A5
subsetc1['S3BQ1A5']=subsetc1['S3BQ1A5'].replace(9, numpy.nan) subsetc2['S3BD5Q2E']=subsetc2['S3BD5Q2E'].replace('BL', numpy.nan) subsetc2['S3BD5Q2E']=subsetc2['S3BD5Q2E'].replace(99, numpy.nan)
Contingency table of observed counts of major depression diagnosis (response variable) within cannabis use (explanatory variable), in ages 18-30
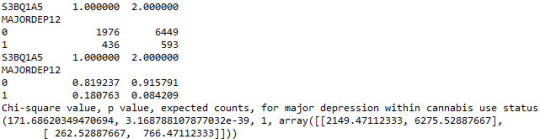
contab1=pandas.crosstab(subsetc1['MAJORDEP12'], subsetc1['S3BQ1A5']) print (contab1)
#Column percentages
colsum=contab1.sum(axis=0) colpcontab=contab1/colsum print(colpcontab)
#Chi-square calculations for major depression within cannabis use status
print ('Chi-square value, p value, expected counts, for major depression within cannabis use status') chsq1= scipy.stats.chi2_contingency(contab1) print (chsq1)
#Contingency table of observed counts of geberal anxiety diagnosis (response variable) within cannabis use (explanatory variable), in ages 18-30
contab2=pandas.crosstab(subsetc1['GENAXDX12'], subsetc1['S3BQ1A5']) print (contab2)
#Column percentages
colsum2=contab2.sum(axis=0) colpcontab2=contab2/colsum2 print(colpcontab2)
#Chi-square calculations for general anxiety within cannabis use status
print ('Chi-square value, p value, expected counts, for general anxiety within cannabis use status') chsq2= scipy.stats.chi2_contingency(contab2) print (chsq2)
#
#Contingency table of observed counts of major depression diagnosis (response variable) within frequency of cannabis use (10 level explanatory variable), in ages 18-30
contab3=pandas.crosstab(subset2['MAJORDEP12'], subset2['S3BD5Q2E']) print (contab3)
#Column percentages
colsum3=contab3.sum(axis=0) colpcontab3=contab3/colsum3 print(colpcontab3)
#Chi-square calculations for mahor depression within frequency of cannabis use groups
print ('Chi-square value, p value, expected counts for major depression associated frequency of cannabis use') chsq3= scipy.stats.chi2_contingency(contab3) print (chsq3)
recode1 = {1: 9, 2: 8, 3: 7, 4: 6, 5: 5, 6: 4, 7: 3, 8: 2, 9: 1} # Dictionary with details of frequency variable reverse-recode subsetc2['CUFREQ'] = subsetc2['S3BD5Q2E'].map(recode1) # Change variable name from S3BD5Q2E to CUFREQ
subsetc2["CUFREQ"] = subsetc2["CUFREQ"].astype('category')
#Rename graph labels for better interpretation
subsetc2['CUFREQ'] = subsetc2['CUFREQ'].cat.rename_categories(["2 times/year","3-6 times/year","7-11 times/years","Once a month","2-3 times/month","1-2 times/week","3-4 times/week","Nearly every day","Every day"])
#Graph percentages of major depression within each cannabis smoking frequency group
plt.figure(figsize=(12,4)) # Change plot size ax1 = seaborn.factorplot(x="CUFREQ", y="MAJORDEP12", data=subsetc2, kind="bar", ci=None) ax1.set_xticklabels(rotation=40, ha="right") # X-axis labels rotation plt.xlabel('Frequency of cannabis use') plt.ylabel('Proportion of Major Depression') plt.show()
#Post hoc test, pair comparison of frequency groups 1 and 9, 'Every day' and '2 times a year'
recode2 = {1: 1, 9: 9} subsetc2['COMP1v9']= subsetc2['S3BD5Q2E'].map(recode2)
#Contingency table of observed counts
ct4=pandas.crosstab(subsetc2['MAJORDEP12'], subsetc2['COMP1v9']) print (ct4)
#Column percentages
colsum4=ct4.sum(axis=0) colpcontab4=ct4/colsum4 print(colpcontab4)
#Chi-square calculations for pair comparison of frequency groups 1 and 9, 'Every day' and '2 times a year'
print ('Chi-square value, p value, expected counts, for pair comparison of frequency groups -Every day- and -2 times a year-') cs4= scipy.stats.chi2_contingency(ct4) print (cs4)
#Post hoc test, pair comparison of frequency groups 2 and 6, 'Nearly every day' and 'Once a month'
recode3 = {2: 2, 6: 6} subsetc2['COMP2v6']= subsetc2['S3BD5Q2E'].map(recode3)
#Contingency table of observed counts
ct5=pandas.crosstab(subsetc2['MAJORDEP12'], subsetc2['COMP2v6']) print (ct5)
#Column percentages
colsum5=ct5.sum(axis=0) colpcontab5=ct5/colsum5 print(colpcontab5)
#Chi-square calculations for pair comparison of frequency groups 2 and 6, 'Nearly every day' and 'Once a month'
print ('Chi-square value, p value, expected counts for pair comparison of frequency groups -Nearly every day- and -Once a month-') cs5= scipy.stats.chi2_contingency(ct5) print (cs5)

1 note
·
View note
Text
Running a Chi-Square Test of Independence for my data
import pandas import numpy import scipy.stats import seaborn import matplotlib.pyplot as plt
nesarc = pandas.read_csv ('nesarc_pds.csv' , low_memory=False)
Set PANDAS to show all columns in DataFrame
pandas.set_option('display.max_columns', None)
#Set PANDAS to show all rows in DataFrame
pandas.set_option('display.max_rows', None)
nesarc.columns = map(str.upper , nesarc.columns)
pandas.set_option('display.float_format' , lambda x:'%f'%x)
#Change my variables to numeric
nesarc['AGE'] = pandas.to_numeric(nesarc['AGE'], errors='coerce') nesarc['S3BQ4'] = pandas.to_numeric(nesarc['S3BQ4'], errors='coerce') nesarc['S3BQ1A5'] = pandas.to_numeric(nesarc['S3BQ1A5'], errors='coerce') nesarc['S3BD5Q2B'] = pandas.to_numeric(nesarc['S3BD5Q2B'], errors='coerce') nesarc['S3BD5Q2E'] = pandas.to_numeric(nesarc['S3BD5Q2E'], errors='coerce') nesarc['MAJORDEP12'] = pandas.to_numeric(nesarc['MAJORDEP12'], errors='coerce') nesarc['GENAXDX12'] = pandas.to_numeric(nesarc['GENAXDX12'], errors='coerce')
#Subset my sample
subset1 = nesarc[(nesarc['AGE']>=18) & (nesarc['AGE']<=30)] # Ages 18-30 subsetc1 = subset1.copy()
subset2 = nesarc[(nesarc['AGE']>=18) & (nesarc['AGE']<=30) & (nesarc['S3BQ1A5']==1)] # Cannabis users, ages 18-30 subsetc2 = subset2.copy()
#Setting missing data for frequency and cannabis use, variables S3BD5Q2E, S3BQ1A5
subsetc1['S3BQ1A5']=subsetc1['S3BQ1A5'].replace(9, numpy.nan) subsetc2['S3BD5Q2E']=subsetc2['S3BD5Q2E'].replace('BL', numpy.nan) subsetc2['S3BD5Q2E']=subsetc2['S3BD5Q2E'].replace(99, numpy.nan)
Contingency table of observed counts of major depression diagnosis (response variable) within cannabis use (explanatory variable), in ages 18-30
contab1=pandas.crosstab(subsetc1['MAJORDEP12'], subsetc1['S3BQ1A5']) print (contab1)
#Column percentages
colsum=contab1.sum(axis=0) colpcontab=contab1/colsum print(colpcontab)
#Chi-square calculations for major depression within cannabis use status
print ('Chi-square value, p value, expected counts, for major depression within cannabis use status') chsq1= scipy.stats.chi2_contingency(contab1) print (chsq1)
#Contingency table of observed counts of geberal anxiety diagnosis (response variable) within cannabis use (explanatory variable), in ages 18-30
contab2=pandas.crosstab(subsetc1['GENAXDX12'], subsetc1['S3BQ1A5']) print (contab2)
#Column percentages
colsum2=contab2.sum(axis=0) colpcontab2=contab2/colsum2 print(colpcontab2)
#Chi-square calculations for general anxiety within cannabis use status
print ('Chi-square value, p value, expected counts, for general anxiety within cannabis use status') chsq2= scipy.stats.chi2_contingency(contab2) print (chsq2)
#
#Contingency table of observed counts of major depression diagnosis (response variable) within frequency of cannabis use (10 level explanatory variable), in ages 18-30
contab3=pandas.crosstab(subset2['MAJORDEP12'], subset2['S3BD5Q2E']) print (contab3)
#Column percentages
colsum3=contab3.sum(axis=0) colpcontab3=contab3/colsum3 print(colpcontab3)
#Chi-square calculations for mahor depression within frequency of cannabis use groups
print ('Chi-square value, p value, expected counts for major depression associated frequency of cannabis use') chsq3= scipy.stats.chi2_contingency(contab3) print (chsq3)
recode1 = {1: 9, 2: 8, 3: 7, 4: 6, 5: 5, 6: 4, 7: 3, 8: 2, 9: 1} # Dictionary with details of frequency variable reverse-recode subsetc2['CUFREQ'] = subsetc2['S3BD5Q2E'].map(recode1) # Change variable name from S3BD5Q2E to CUFREQ
subsetc2["CUFREQ"] = subsetc2["CUFREQ"].astype('category')
#Rename graph labels for better interpretation
subsetc2['CUFREQ'] = subsetc2['CUFREQ'].cat.rename_categories(["2 times/year","3-6 times/year","7-11 times/years","Once a month","2-3 times/month","1-2 times/week","3-4 times/week","Nearly every day","Every day"])
#Graph percentages of major depression within each cannabis smoking frequency group
plt.figure(figsize=(12,4)) # Change plot size ax1 = seaborn.factorplot(x="CUFREQ", y="MAJORDEP12", data=subsetc2, kind="bar", ci=None) ax1.set_xticklabels(rotation=40, ha="right") # X-axis labels rotation plt.xlabel('Frequency of cannabis use') plt.ylabel('Proportion of Major Depression') plt.show()
#Post hoc test, pair comparison of frequency groups 1 and 9, 'Every day' and '2 times a year'
recode2 = {1: 1, 9: 9} subsetc2['COMP1v9']= subsetc2['S3BD5Q2E'].map(recode2)
#Contingency table of observed counts
ct4=pandas.crosstab(subsetc2['MAJORDEP12'], subsetc2['COMP1v9']) print (ct4)
#Column percentages
colsum4=ct4.sum(axis=0) colpcontab4=ct4/colsum4 print(colpcontab4)
#Chi-square calculations for pair comparison of frequency groups 1 and 9, 'Every day' and '2 times a year'
print ('Chi-square value, p value, expected counts, for pair comparison of frequency groups -Every day- and -2 times a year-') cs4= scipy.stats.chi2_contingency(ct4) print (cs4)
#Post hoc test, pair comparison of frequency groups 2 and 6, 'Nearly every day' and 'Once a month'
recode3 = {2: 2, 6: 6} subsetc2['COMP2v6']= subsetc2['S3BD5Q2E'].map(recode3)
#Contingency table of observed counts
ct5=pandas.crosstab(subsetc2['MAJORDEP12'], subsetc2['COMP2v6']) print (ct5)
#Column percentages
colsum5=ct5.sum(axis=0) colpcontab5=ct5/colsum5 print(colpcontab5)
#Chi-square calculations for pair comparison of frequency groups 2 and 6, 'Nearly every day' and 'Once a month'
print ('Chi-square value, p value, expected counts for pair comparison of frequency groups -Nearly every day- and -Once a month-') cs5= scipy.stats.chi2_contingency(ct5) print (cs5)
0 notes
Text
Pandas
To import Pandas; import pandas as pd
To create a dataframe from a csv: fd = pd.read_csv("title.csv")
To see the first 5 rows of a dataframe: df.head()
To get the number of rows and columns: df.shape
To get the names of the columns: df.columns
To see NaN (not a number) values (where True = NaN): df.isna()
To see the last 5 rows of a dataframe: df.tail()
To create a clean dataframe without rows with NaN: clean_df = df.dropna()
To access a particular column by name: clean_df['Starting Median Salary']
To find the highest value in a column: clean_df['Starting Median Salary'].max()
To get the row number or index of that value: clean_df['Starting Median Salary'].idxmax()
To get the value from another column at that index: clean_df['Undergraduate Major'].loc[43] OR clean_df['Undergraduate Major'][43]
To get the entire row at a given index: clean_df.loc[43]
To get the difference between two columns:
clean_df['Mid-Career 90th Percentile Salary'] - clean_df['Mid-Career 10th Percentile Salary'] OR
clean_df['Mid-Career 90th Percentile Salary'].subtract(clean_df['Mid-Career 10th Percentile Salary'])
To insert this as a new column;
spread_col = clean_df['Mid-Career 90th Percentile Salary'] - clean_df['Mid-Career 10th Percentile Salary']
clean_df.insert(1, 'Spread', spread_col)
clean_df.head()
To create a new table sorted by a column: low_risk = clean_df.sort_values('Spread')
To only display two columns: low_risk[['Undergraduate Major', 'Spread']].head()
To see how many of each type you have:
clean_df.groupby('Group').sum()
To count how many you have by of each category: clean_df.groupby('Group').count()
To round to two decimal places:
pd.options.display.float_format = '{:,.2f}'.format
To get the averages for each category:
clean_df.groupby('Group').mean()
To rename columns:
df = pd.read_csv('QueryResults.csv', names=['DATE', 'TAG', 'POSTS'], header=0)
To get the sum of entries:
df.groupby("TAG").sum()
To count how many entries there are:
df.groupby("TAG").count()
To select an individual cell:
df['DATE'][1]
or df.DATE[1]
To inspect the datatype:
type(df["DATE"][1])
To convert a string into a datetime:
df.DATE = pd.to_datetime(df.DATE)
To pivot a dataframe:
reshaped_df = df.pivot(index='DATE', columns='TAG', values='POSTS')
To replace NaN with zeros:
reshaped_df.fillna(0, inplace=True) or
reshaped_df = reshaped_df.fillna(0)
To check there aren't any NaN values left:
reshaped_df.isna().values.any()
To count how many of each type there is:
colors.groupby("is_trans").count() or
colors.is_trans.value_counts()
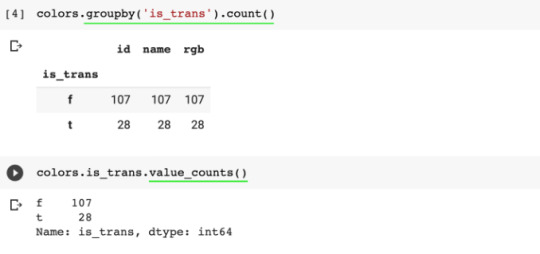
To find all the entries with a certain value (to filter by a condition):
sets[sets['year'] == 1949]
To aggregate data:
themes_by_year = sets.groupby('year').agg({'theme_id': pd.Series.nunique})
Note, the .agg() method takes a dictionary as an argument. In this dictionary, we specify which operation we'd like to apply to each column. In our case, we just want to calculate the number of unique entries in the theme_id column by using our old friend, the .nunique() method.
To rename columns:
themes_by_year.rename(columns = {'theme_id': 'nr_themes'}, inplace= True)
To plot:
plt.plot(themes_by_year.index[:-2], themes_by_year.nr_themes[:-2])
To plot two lines with two axis:
ax1 = plt.gca() # get current axes
ax2 = ax1.twinx() #allows them to share the same x-axis
ax1.plot(themes_by_year.index[:-2], themes_by_year.nr_themes[:-2])
ax2.plot(sets_by_year.index[:-2], sets_by_year.set_num[:-2])
ax1.set_xlabel("Year")
ax1.set_ylabel("Number of Sets", color="green")
ax2.set_ylabel("Number of Themes", color="blue")
To get the average number of parts per year:
parts_per_set = sets.groupby('year').agg({'num_parts': pd.Series.mean})
To change daily data to monthly data:
df_btc_monthly.head()
0 notes
Text
Running a k-means Cluster Analysis
from pandas import Series, DataFrame
import pandas as pd
import numpy as np
import os
import matplotlib.pylab as plt
from sklearn.cross_validation import train_test_split
from sklearn.model_selection import train_test_split
from sklearn import preprocessing
from sklearn.cluster import KMeans
"""
Data Management
"""
os.chdir("C:\TREES")
data = pd.read_csv("tree_addhealth.csv")
upper-case all DataFrame column names
data.columns = map(str.upper, data.columns)
Data Management
data_clean = data.dropna()
subset clustering variables
cluster=data_clean[['ALCEVR1','MAREVER1','ALCPROBS1','DEVIANT1','VIOL1',
'DEP1','ESTEEM1','SCHCONN1','PARACTV', 'PARPRES','FAMCONCT']]
cluster.describe()
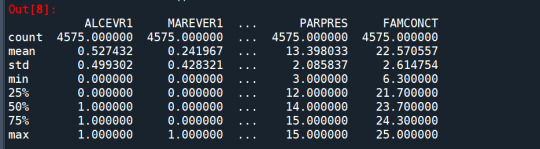
standardize clustering variables to have mean=0 and sd=1
clustervar=cluster.copy()
clustervar['ALCEVR1']=preprocessing.scale(clustervar['ALCEVR1'].astype('float64'))
clustervar['ALCPROBS1']=preprocessing.scale(clustervar['ALCPROBS1'].astype('float64'))
clustervar['MAREVER1']=preprocessing.scale(clustervar['MAREVER1'].astype('float64'))
clustervar['DEP1']=preprocessing.scale(clustervar['DEP1'].astype('float64'))
clustervar['ESTEEM1']=preprocessing.scale(clustervar['ESTEEM1'].astype('float64'))
clustervar['VIOL1']=preprocessing.scale(clustervar['VIOL1'].astype('float64'))
clustervar['DEVIANT1']=preprocessing.scale(clustervar['DEVIANT1'].astype('float64'))
clustervar['FAMCONCT']=preprocessing.scale(clustervar['FAMCONCT'].astype('float64'))
clustervar['SCHCONN1']=preprocessing.scale(clustervar['SCHCONN1'].astype('float64'))
clustervar['PARACTV']=preprocessing.scale(clustervar['PARACTV'].astype('float64'))
clustervar['PARPRES']=preprocessing.scale(clustervar['PARPRES'].astype('float64'))
split data into train and test sets
clus_train, clus_test = train_test_split(clustervar, test_size=.3, random_state=123)
k-means cluster analysis for 1-9 clusters
from scipy.spatial.distance import cdist
clusters=range(1,10)
meandist=[]
for k in clusters:
model=KMeans(n_clusters=k)
model.fit(clus_train)
clusassign=model.predict(clus_train)
meandist.append(sum(np.min(cdist(clus_train, model.cluster_centers_, 'euclidean'), axis=1))
/ clus_train.shape[0])
"""
Plot average distance from observations from the cluster centroid
to use the Elbow Method to identify number of clusters to choose
"""
plt.plot(clusters, meandist)
plt.xlabel('Number of clusters')
plt.ylabel('Average distance')
plt.title('Selecting k with the Elbow Method')
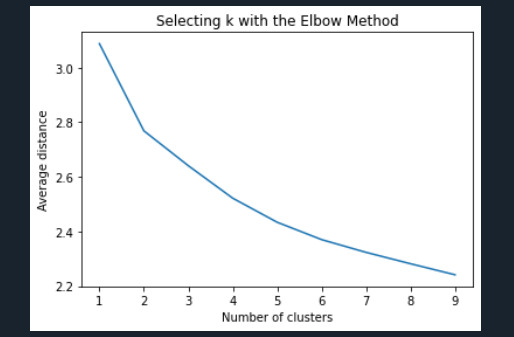
Interpret 3 cluster solution
model3=KMeans(n_clusters=3)
model3.fit(clus_train)
clusassign=model3.predict(clus_train)
plot clusters
from sklearn.decomposition import PCA
pca_2 = PCA(2)
plot_columns = pca_2.fit_transform(clus_train)
plt.scatter(x=plot_columns[:,0], y=plot_columns[:,1], c=model3.labels_,)
plt.xlabel('Canonical variable 1')
plt.ylabel('Canonical variable 2')
plt.title('Scatterplot of Canonical Variables for 3 Clusters')
plt.show()
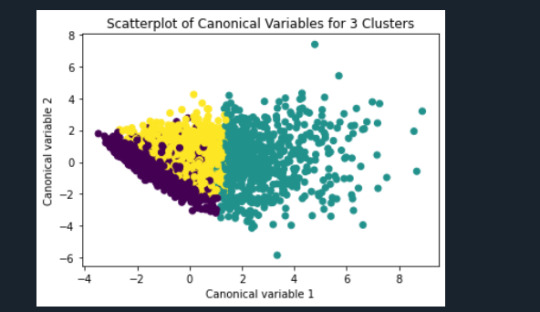
"""
BEGIN multiple steps to merge cluster assignment with clustering variables to examine
cluster variable means by cluster
"""
create a unique identifier variable from the index for the
cluster training data to merge with the cluster assignment variable
clus_train.reset_index(level=0, inplace=True)
create a list that has the new index variable
cluslist=list(clus_train['index'])
create a list of cluster assignments
labels=list(model3.labels_)
combine index variable list with cluster assignment list into a dictionary
newlist=dict(zip(cluslist, labels))
newlist
convert newlist dictionary to a dataframe
newclus=DataFrame.from_dict(newlist, orient='index')
newclus
rename the cluster assignment column
newclus.columns = ['cluster']
now do the same for the cluster assignment variable
create a unique identifier variable from the index for the
cluster assignment dataframe
to merge with cluster training data
newclus.reset_index(level=0, inplace=True)
merge the cluster assignment dataframe with the cluster training variable dataframe
by the index variable
merged_train=pd.merge(clus_train, newclus, on='index')
merged_train.head(n=100)
cluster frequencies
merged_train.cluster.value_counts()
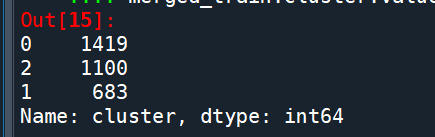
"""
END multiple steps to merge cluster assignment with clustering variables to examine
cluster variable means by cluster
"""
FINALLY calculate clustering variable means by cluster
clustergrp = merged_train.groupby('cluster').mean()
print ("Clustering variable means by cluster")
print(clustergrp)
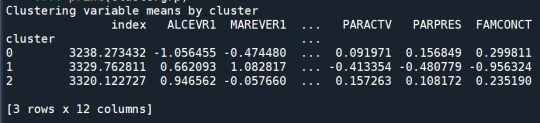
validate clusters in training data by examining cluster differences in GPA using ANOVA
first have to merge GPA with clustering variables and cluster assignment data
gpa_data=data_clean['GPA1']
split GPA data into train and test sets
gpa_train, gpa_test = train_test_split(gpa_data, test_size=.3, random_state=123)
gpa_train1=pd.DataFrame(gpa_train)
gpa_train1.reset_index(level=0, inplace=True)
merged_train_all=pd.merge(gpa_train1, merged_train, on='index')
sub1 = merged_train_all[['GPA1', 'cluster']].dropna()
import statsmodels.formula.api as smf
import statsmodels.stats.multicomp as multi
gpamod = smf.ols(formula='GPA1 ~ C(cluster)', data=sub1).fit()
print (gpamod.summary())
print ('means for GPA by cluster')
m1= sub1.groupby('cluster').mean()
print (m1)
print ('standard deviations for GPA by cluster')
m2= sub1.groupby('cluster').std()
print (m2)
mc1 = multi.MultiComparison(sub1['GPA1'], sub1['cluster'])
res1 = mc1.tukeyhsd()
print(res1.summary())
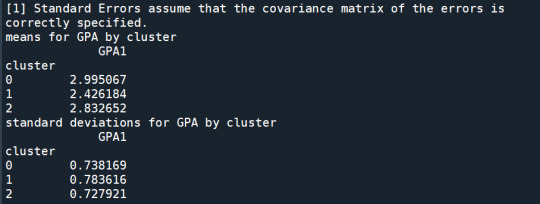
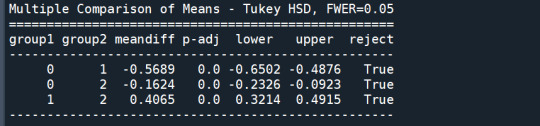
In order to externally validate the clusters, an Analysis of Variance (ANOVA) was conducting to test for significant differences between the clusters on grade point average (GPA). A tukey test was used for post hoc comparisons between the clusters. Results indicated significant differences between the clusters on GPA (F(3, 3197)=82.28, p<.0001). The tukey post hoc comparisons showed significant differences between clusters on GPA, with the exception that clusters 1 and 2 were not significantly different from each other. Adolescents in cluster 4 had the highest GPA (mean=2.99, sd=0.73), and cluster 3 had the lowest GPA (mean=2.42, sd=0.78).
0 notes
Text
Running a Chi-Square Test of Independence
import pandas
import numpy
import scipy.stats
import seaborn
import matplotlib.pyplot as plt
nesarc = pandas.read_csv ('nesarc_pds.csv' , low_memory=False)
Set PANDAS to show all columns in DataFrame
pandas.set_option('display.max_columns', None)
Set PANDAS to show all rows in DataFrame
pandas.set_option('display.max_rows', None)
nesarc.columns = map(str.upper , nesarc.columns)
pandas.set_option('display.float_format' , lambda x:'%f'%x)
Change my variables to numeric
nesarc['AGE'] = pandas.to_numeric(nesarc['AGE'], errors='coerce')
nesarc['S3BQ4'] = pandas.to_numeric(nesarc['S3BQ4'], errors='coerce')
nesarc['S3BQ1A5'] = pandas.to_numeric(nesarc['S3BQ1A5'], errors='coerce')
nesarc['S3BD5Q2B'] = pandas.to_numeric(nesarc['S3BD5Q2B'], errors='coerce')
nesarc['S3BD5Q2E'] = pandas.to_numeric(nesarc['S3BD5Q2E'], errors='coerce')
nesarc['MAJORDEP12'] = pandas.to_numeric(nesarc['MAJORDEP12'], errors='coerce')
nesarc['GENAXDX12'] = pandas.to_numeric(nesarc['GENAXDX12'], errors='coerce')
Subset my sample
subset1 = nesarc[(nesarc['AGE']>=18) & (nesarc['AGE']<=30)] # Ages 18-30
subsetc1 = subset1.copy()
subset2 = nesarc[(nesarc['AGE']>=18) & (nesarc['AGE']<=30) & (nesarc['S3BQ1A5']==1)] # Cannabis users, ages 18-30
subsetc2 = subset2.copy()
Setting missing data for frequency and cannabis use, variables S3BD5Q2E, S3BQ1A5
subsetc1['S3BQ1A5']=subsetc1['S3BQ1A5'].replace(9, numpy.nan)
subsetc2['S3BD5Q2E']=subsetc2['S3BD5Q2E'].replace('BL', numpy.nan)
subsetc2['S3BD5Q2E']=subsetc2['S3BD5Q2E'].replace(99, numpy.nan)
Contingency table of observed counts of major depression diagnosis (response variable) within cannabis use (explanatory variable), in ages 18-30
contab1=pandas.crosstab(subsetc1['MAJORDEP12'], subsetc1['S3BQ1A5'])
print (contab1)
Column percentages
colsum=contab1.sum(axis=0)
colpcontab=contab1/colsum
print(colpcontab)
Chi-square calculations for major depression within cannabis use status
print ('Chi-square value, p value, expected counts, for major depression within cannabis use status')
chsq1= scipy.stats.chi2_contingency(contab1)
print (chsq1)
Contingency table of observed counts of geberal anxiety diagnosis (response variable) within cannabis use (explanatory variable), in ages 18-30
contab2=pandas.crosstab(subsetc1['GENAXDX12'], subsetc1['S3BQ1A5'])
print (contab2)
Column percentages
colsum2=contab2.sum(axis=0)
colpcontab2=contab2/colsum2
print(colpcontab2)
Chi-square calculations for general anxiety within cannabis use status
print ('Chi-square value, p value, expected counts, for general anxiety within cannabis use status')
chsq2= scipy.stats.chi2_contingency(contab2)
print (chsq2)
#
Contingency table of observed counts of major depression diagnosis (response variable) within frequency of cannabis use (10 level explanatory variable), in ages 18-30
contab3=pandas.crosstab(subset2['MAJORDEP12'], subset2['S3BD5Q2E'])
print (contab3)
Column percentages
colsum3=contab3.sum(axis=0)
colpcontab3=contab3/colsum3
print(colpcontab3)
Chi-square calculations for mahor depression within frequency of cannabis use groups
print ('Chi-square value, p value, expected counts for major depression associated frequency of cannabis use')
chsq3= scipy.stats.chi2_contingency(contab3)
print (chsq3)
recode1 = {1: 9, 2: 8, 3: 7, 4: 6, 5: 5, 6: 4, 7: 3, 8: 2, 9: 1} # Dictionary with details of frequency variable reverse-recode
subsetc2['CUFREQ'] = subsetc2['S3BD5Q2E'].map(recode1) # Change variable name from S3BD5Q2E to CUFREQ
subsetc2["CUFREQ"] = subsetc2["CUFREQ"].astype('category')
Rename graph labels for better interpretation
subsetc2['CUFREQ'] = subsetc2['CUFREQ'].cat.rename_categories(["2 times/year","3-6 times/year","7-11 times/years","Once a month","2-3 times/month","1-2 times/week","3-4 times/week","Nearly every day","Every day"])
Graph percentages of major depression within each cannabis smoking frequency group
plt.figure(figsize=(12,4)) # Change plot size
ax1 = seaborn.factorplot(x="CUFREQ", y="MAJORDEP12", data=subsetc2, kind="bar", ci=None)
ax1.set_xticklabels(rotation=40, ha="right") # X-axis labels rotation
plt.xlabel('Frequency of cannabis use')
plt.ylabel('Proportion of Major Depression')
plt.show()
Post hoc test, pair comparison of frequency groups 1 and 9, 'Every day' and '2 times a year'
recode2 = {1: 1, 9: 9}
subsetc2['COMP1v9']= subsetc2['S3BD5Q2E'].map(recode2)
Contingency table of observed counts
ct4=pandas.crosstab(subsetc2['MAJORDEP12'], subsetc2['COMP1v9'])
print (ct4)
Column percentages
colsum4=ct4.sum(axis=0)
colpcontab4=ct4/colsum4
print(colpcontab4)
Chi-square calculations for pair comparison of frequency groups 1 and 9, 'Every day' and '2 times a year'
print ('Chi-square value, p value, expected counts, for pair comparison of frequency groups -Every day- and -2 times a year-')
cs4= scipy.stats.chi2_contingency(ct4)
print (cs4)
Post hoc test, pair comparison of frequency groups 2 and 6, 'Nearly every day' and 'Once a month'
recode3 = {2: 2, 6: 6}
subsetc2['COMP2v6']= subsetc2['S3BD5Q2E'].map(recode3)
Contingency table of observed counts
ct5=pandas.crosstab(subsetc2['MAJORDEP12'], subsetc2['COMP2v6'])
print (ct5)
Column percentages
colsum5=ct5.sum(axis=0)
colpcontab5=ct5/colsum5
print(colpcontab5)
Chi-square calculations for pair comparison of frequency groups 2 and 6, 'Nearly every day' and 'Once a month'
print ('Chi-square value, p value, expected counts for pair comparison of frequency groups -Nearly every day- and -Once a month-')
cs5= scipy.stats.chi2_contingency(ct5)
print (cs5)
OUTPUT
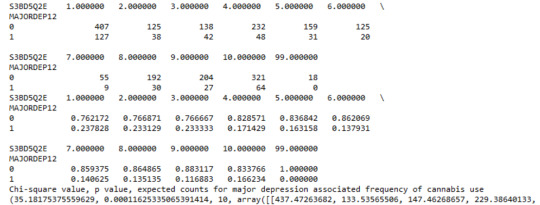
A Chi Square test of independence revealed that among cannabis users aged between 18 and 30 years old (subsetc2), the frequency of cannabis use (explanatory variable collapsed into 10 ordered categories) and past year depression diagnosis (response binary categorical variable) were significantly associated, X2 =35.18, 10 df, p=0.00011.
Similarly, the post hoc comparison (Bonferroni Adjustment) of rates of major depression by the pair of "Nearly every day” and “once a month” frequency categories, indicated that the p-value is 0.046 and the proportions of major depression diagnosis for each frequency group are 23.3% and 13.7% respectively. As a result, since the p-value is larger than the Bonferroni adjusted p-value (adj p-value = 0.05 / 45 = 0.0011<0.046), we can assume that these two rates are not significantly different from one another. Therefore, we accept the null hypothesis.
0 notes
Text
Chi-Square Test
Week2: Running a Chi-Square Test of Independence
import pandas
import numpy
import scipy.stats
import seaborn
import matplotlib.pyplot as plt
NESARC Dataset
nesarc = pandas.read_csv ('nesarc_pds.csv' , low_memory=False)
#Show all columns in DataFrame
pandas.set_option('display.max_columns', None)
#Show all rows in DataFrame
pandas.set_option('display.max_rows', None)
nesarc.columns = map(str.upper , nesarc.columns)
pandas.set_option('display.float_format' , lambda x:'%f'%x)
# Convert variables to numeric
nesarc['AGE'] = pandas.to_numeric(nesarc['AGE'], errors='coerce')
nesarc['S3BQ4'] = pandas.to_numeric(nesarc['S3BQ4'], errors='coerce')
nesarc['S3BQ1A5'] = pandas.to_numeric(nesarc['S3BQ1A5'], errors='coerce')
nesarc['S3BD5Q2B'] = pandas.to_numeric(nesarc['S3BD5Q2B'], errors='coerce')
nesarc['S3BD5Q2E'] = pandas.to_numeric(nesarc['S3BD5Q2E'], errors='coerce')
nesarc['MAJORDEP12'] = pandas.to_numeric(nesarc['MAJORDEP12'], errors='coerce')
nesarc['GENAXDX12'] = pandas.to_numeric(nesarc['GENAXDX12'], errors='coerce')
# Data for Ages 18-30
subset1 = nesarc[(nesarc['AGE']>=18) & (nesarc['AGE']<=30)]
subsetc1 = subset1.copy()
# Cannabis users, ages 18-30
subset2 = nesarc[(nesarc['AGE']>=18) & (nesarc['AGE']<=30) & (nesarc['S3BQ1A5']==1)]
subsetc2 = subset2.copy()
# Setting missing data for frequency and cannabis use, variables S3BD5Q2E, S3BQ1A5
subsetc1['S3BQ1A5']=subsetc1['S3BQ1A5'].replace(9, numpy.nan)
subsetc2['S3BD5Q2E']=subsetc2['S3BD5Q2E'].replace('BL', numpy.nan)
subsetc2['S3BD5Q2E']=subsetc2['S3BD5Q2E'].replace(99, numpy.nan)
# Contingency table of observed counts of major depression diagnosis
(response variable) within cannabis use (explanatory variable), in ages 18-30
contab1=pandas.crosstab(subsetc1['MAJORDEP12'], subsetc1['S3BQ1A5'])
print (contab1)
# Column percentages
colsum=contab1.sum(axis=0)
colpcontab=contab1/colsum
print(colpcontab)
# Chi-square calculations for major depression within cannabis use status
print ('Chi-square value, p value, expected counts, for major depression within cannabis use status')
chsq1= scipy.stats.chi2_contingency(contab1)
print (chsq1)
# Contingency table of observed counts of general anxiety diagnosis(response variable) within cannabis use (explanatory variable), in ages 18-30
contab2=pandas.crosstab(subsetc1['GENAXDX12'], subsetc1['S3BQ1A5'])
print (contab2)
# Column percentages
colsum2=contab2.sum(axis=0)
colpcontab2=contab2/colsum2
print(colpcontab2)
# Chi-square calculations for general anxiety within cannabis use status
print ('Chi-square value, p value, expected counts, for general anxiety within cannabis use status')
chsq2= scipy.stats.chi2_contingency(contab2)
print (chsq2)
# Contingency table for observed counts of major depression diagnosis(response variable) and frequency of cannabis use (10 level explanatory variable), between age 18-30
contab3=pandas.crosstab(subset2['MAJORDEP12'], subset2['S3BD5Q2E'])
print (contab3)
# Column percentages
colsum3=contab3.sum(axis=0)
colpcontab3=contab3/colsum3
print(colpcontab3)
# Chi-square calculations for major depression within frequency of cannabis use groups
print ('Chi-square value, p value, expected counts for major depression associated frequency of cannabis use')
chsq3= scipy.stats.chi2_contingency(contab3)
print (chsq3)
# Dictionary with details of frequency variable reverse-recode
recode1 = {1: 9, 2: 8, 3: 7, 4: 6, 5: 5, 6: 4, 7: 3, 8: 2, 9: 1}
# Change variable name from S3BD5Q2E to CUFREQ
subsetc2['CUFREQ'] = subsetc2['S3BD5Q2E'].map(recode1)
subsetc2["CUFREQ"] = subsetc2["CUFREQ"].astype('category')
# Rename graph labels for better interpretation
subsetc2['CUFREQ'] = subsetc2['CUFREQ'].cat.rename_categories(["2 times/year","3-6 times/year","7-11 times/years","Once a month","2-3 times/month","1-2 times/week","3-4 times/week","Nearly every day","Every day"])
# Graph percentages of major depression within each cannabis smoking frequency group
plt.figure(figsize=(12,4)) # Change plot size
ax1 = seaborn.factorplot(x="CUFREQ", y="MAJORDEP12", data=subsetc2, kind="bar", ci=None)
ax1.set_xticklabels(rotation=40, ha="right") # X-axis labels rotation
plt.xlabel('Frequency of cannabis use')
plt.ylabel('Proportion of Major Depression')
plt.show()
# Post hoc test, pair comparison of frequency groups 1 and 9, 'Every day' and '2 times a year'
recode2 = {1: 1, 9: 9}
subsetc2['COMP1v9']= subsetc2['S3BD5Q2E'].map(recode2)
# Contingency table of observed counts
ct4=pandas.crosstab(subsetc2['MAJORDEP12'], subsetc2['COMP1v9'])
print (ct4)
# Column percentages
colsum4=ct4.sum(axis=0)
colpcontab4=ct4/colsum4
print(colpcontab4)
# Chi-square calculations for pair comparison of frequency groups 1 and 9, 'Every day' and '2 times a year'
print ('Chi-square value, p value, expected counts, for pair comparison of frequency groups -Every day- and -2 times a year-')
cs4= scipy.stats.chi2_contingency(ct4)
print (cs4)
# Post hoc test, pair comparison of frequency groups 2 and 6, 'Nearly every day' and 'Once a month'
recode3 = {2: 2, 6: 6}
subsetc2['COMP2v6']= subsetc2['S3BD5Q2E'].map(recode3)
# Contingency table of observed counts
ct5=pandas.crosstab(subsetc2['MAJORDEP12'], subsetc2['COMP2v6'])
print (ct5)
# Column percentages
colsum5=ct5.sum(axis=0)
colpcontab5=ct5/colsum5
print(colpcontab5)
# Chi-square calculations for pair comparison of frequency groups 2 and 6, 'Nearly every day' and 'Once a month'
print ('Chi-square value, p value, expected counts for pair comparison of frequency groups -Nearly every day- and -Once a month-')
cs5= scipy.stats.chi2_contingency(ct5)
print (cs5)
Model Interpretation for Chi-Square Tests:
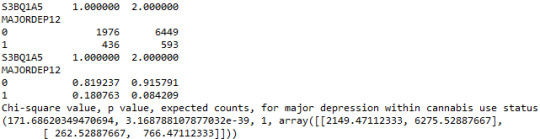
When examining the patterns of association between major depression (categorical response variable) and cannabis use status (categorical explanatory variable), a chi-square test of independence revealed that among young adults aged between 18 and 30 years old (subsetc1), those who were cannabis users, were more likely to have been diagnosed with major depression in the last 12 months (18%), compared to the non-users (8.4%), X2 =171.6, 1 df, p=3.16e-39 (p-value is written in scientific notation). As a result, since our p-value is extremely small, the data provides significant evidence against the null hypothesis. Thus, we reject the null hypothesis and accept the alternate hypothesis, which indicates that there is a positive correlation between cannabis use and depression diagnosis.
Output
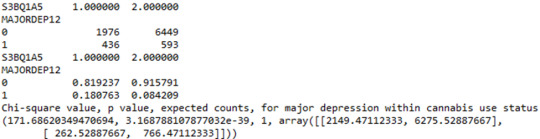
When examining the patterns of association between major depression (categorical response variable) and cannabis use status (categorical explanatory variable), a chi-square test of independence revealed that among young adults aged between 18 and 30 years old (subsetc1), those who were cannabis users, were more likely to have been diagnosed with major depression in the last 12 months (18%), compared to the non-users (8.4%), X2 =171.6, 1 df, p=3.16e-39 (p-value is written in scientific notation). As a result, since our p-value is extremely small, the data provides significant evidence against the null hypothesis. Thus, we reject the null hypothesis and accept the alternate hypothesis, which indicates that there is a positive correlation between cannabis use and depression diagnosis.
Output
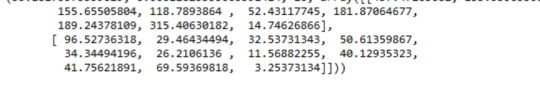
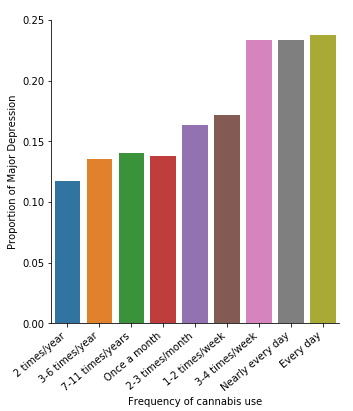
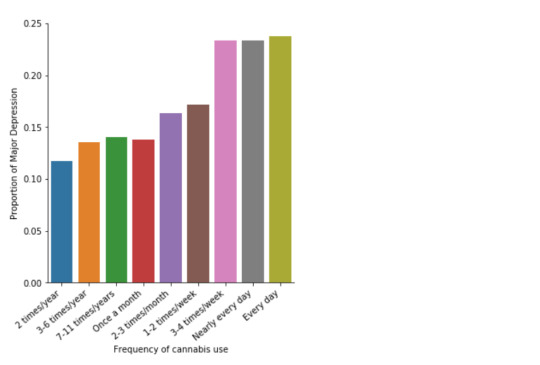
A Chi Square test of independence revealed that among cannabis users aged between 18 and 30 years old (subsetc2), the frequency of cannabis use (explanatory variable collapsed into 10 ordered categories) and past year depression diagnosis (response binary categorical variable) were significantly associated, X2 =35.18, 10 df, p=0.00011.
In the bivariate graph (C->C) below, there is a correlation between frequency of cannabis use (explanatory variable) and major depression diagnosis in the past year (response variable). A left-skewed distribution, indicates that the more an individual aged between 18-30 smoked cannabis, the better the chances to have experienced depression in the last 12 months.
Model Interpretation for post hoc Chi-Square Test results:
The post hoc comparison (Bonferroni Adjustment) of rates of major depression by the pair of “Every day” and “2 times a year” frequency categories, revealed that the p-value is 0.00019 and the percentages of major depression diagnosis for each frequency group are 23.7% and 11.6% respectively. As a result, since the p-value is smaller than the Bonferroni adjusted p-value (adj p-value = 0.05 / 45 = 0.0011>0.00019), it can be assumed that these two rates are significantly different from one another. Therefore, we reject the null hypothesis and accept the alternate.
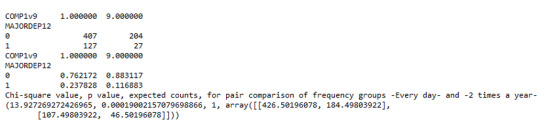
Similarly, the post hoc comparison (Bonferroni Adjustment) of rates of major depression by the pair of "Nearly every day” and “once a month” frequency categories, indicated that the p-value is 0.046 and the proportions of major depression diagnosis for each frequency group are 23.3% and 13.7% respectively. As a result, since the p-value is larger than the Bonferroni adjusted p-value (adj p-value = 0.05 / 45 = 0.0011<0.046), it can be assumed that these two rates are not significantly different from one another. Therefore, we accept the null hypothesis.
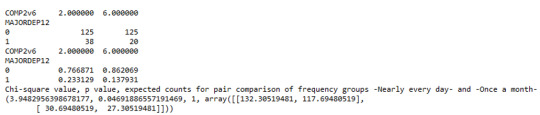
0 notes
Text
Changing a specific column name in pandas DataFrame [duplicate]
This question already has answers here:
Renaming column names in Pandas (32 answers)
Closed 3 months ago.
I was looking for an elegant way to change a specified column name in a DataFrame.
play data ...
import pandas as pdd = { 'one': [1, 2, 3, 4, 5], 'two': [9, 8, 7, 6, 5], 'three': ['a', 'b', 'c', 'd', 'e'] }df = pd.DataFrame(d)
The most elegant solution I have found so far ...
names = df.columns.tolist()names[names.index('two')] = 'new_name'df.columns = names
I was hoping for a simple one-liner ... this attempt failed ...
df.columns[df.columns.tolist().index('one')] = 'another_name'
Any hints gratefully received.
https://codehunter.cc/a/python/changing-a-specific-column-name-in-pandas-dataframe-duplicate
0 notes